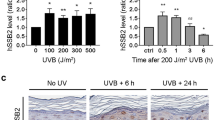
Abstract
The function of human XPA protein, a key subunit of the nucleotide excision repair pathway, has been examined with site-directed substitutions in its putative DNA-binding cleft. After screening for repair activity in a host-cell reactivation assay, we analyzed mutants by comparing their affinities for different substrate architectures, including DNA junctions that provide a surrogate for distorted reaction intermediates, and by testing their ability to recruit the downstream endonuclease partner. Normal repair proficiency was retained when XPA mutations abolished only the simple interaction with linear DNA molecules. By contrast, results from a K141E K179E double mutant revealed that excision is crucially dependent on the assembly of XPA protein with a sharp bending angle in the DNA substrate. These findings show how an increased deformability of damaged sites, leading to helical kinks recognized by XPA, contributes to target selectivity in DNA repair.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sancar, A. DNA excision repair. Annu. Rev. Biochem. 65, 43–81 (1996).
Wood, R.D. Nucleotide excision repair in mammalian cells. J. Biol. Chem. 272, 23465–23468 (1997).
De Laat, W.L., Jaspers, N.G. & Hoeijmakers, J.H. Molecular mechanism of nucleotide excision repair. Genes Dev. 13, 768–785 (1999).
Mitchell, D.L. The induction and repair of lesions produced by the photolysis of (6–4) photoproducts in normal and UV-hypersensitive human cells. Mutat. Res. 194, 227–237 (1988).
Reardon, J.T. & Sancar, A. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 17, 2539–2551 (2003).
Friedberg, E.C., Walker, G.C. & Siede, W. DNA Repair and Mutagenesis (ASM Press, Washington, DC, USA, 1995).
Kraemer, K.H., Lee, M.M. & Scotto, J. DNA repair protects against cutaneous and internal neoplasia: evidence from xeroderma pigmentosum. Carcinogenesis 5, 511–514 (1984).
States, J.C., McDuffie, E.R., Myrand, S.P., McDowell, M. & Cleaver, J.E. Distribution of mutations in the human xeroderma pigementosum group A gene and their relationships to the functional regions of the DNA damage recognition protein. Hum. Mutat. 12, 103–113 (1998).
Cleaver, J.E., Thompson, L.H., Richardson, A.S. & States, J.C. A summary of mutations in the UV-sensitive disorders: xeroderma pigementosum, Cockayne syndrome, and trichothiodystrophy. Hum. Mutat. 14, 9–22 (1999).
Huang, J.C., Svoboda, D., Reardon, J.T. & Sancar, A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl. Acad. Sci. USA 89, 3664–3668 (1992).
Evans, E., Fellows, J., Coffer, A. & Wood, R.D. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 16, 625–638 (1997).
Aboussekhra, A. et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80, 859–868 (1995).
Mellon, I., Spivak, G. & Hanawalt, P.C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51, 241–249 (1987).
Friedberg, E.C. DNA damage and repair. Nature 421, 436–440 (2003).
Mu, D. et al. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270, 2415–2418 (1995).
Sugasawa, K. et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2, 223–232 (1998).
Araújo, S.J. et al. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 14, 349–359 (2000).
Volker, M. et al. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8, 213–224 (2001).
Yang, Z.G., Liu, Y., Mao, L.Y., Zhang, J.-T. & Zou, Y. Dimerization of human XPA and formation of XPA2-RPA protein complex. Biochemistry 41, 13012–13020 (2002).
He, Z., Henricksen, L.A., Wold, M.S. & Ingles, C.J. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature 374, 566–569 (1995).
Li, L., Lu, X., Peterson, C.A. & Legerski, R.J. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol. Cell. Biol. 15, 5396–5402 (1995).
Park, C.H., Mu, D., Reardon, J.T. & Sancar, A. The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J. Biol. Chem. 270, 4896–4902 (1995).
Kuraoka, I. et al. Identification of a damaged-DNA binding domain of the XPA protein. Mutat. Res. 362, 87–95 (1996).
Cleaver, J.E. & States, J.C. The DNA damage-recognition problem in human and other eukaryotic cells: the XPA damage binding protein. Biochem. J. 328, 1–12 (1997).
Ikegami, T. et al. Solution structure of the DNA-and RPA-binding domain of the human repair factor XPA. Nat. Struct. Biol. 5, 701–706 (1998).
Buchko, G.W. et al. DNA-XPA interactions: a 31P NMR and molecular modeling study of dCCAATAACC association with the minimal DNA-binding domain (M98–F219) of the nucleotide excision repair protein XPA. Nucleic Acids Res. 29, 2635–2643 (2001).
Carreau, M. et al. Development of a new easy complementation assay for DNA repair deficient human syndromes using cloned repair genes. Carcinogenesis 16, 1003–1009 (1995).
Mellon, I., Hock, T., Reid, R., Porter, P.C. & States, J.C. Polymorphisms in the human xeroderma pigmentosum group A gene and their impact on cell survival and nucleotide excision repair. DNA Repair (Amst.) 1, 531–546 (2002).
Jones, C.J. & Wood, R.D. Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry 32, 12096–12104 (1993).
You, J.S., Wang, M. & Lee, S.H. Biochemical analysis of the damage recognition process in nucleotide excision repair. J. Biol. Chem. 278, 7476–7485 (2003).
Werner, M.H., Gronenborn, A.M. & Clore, G.M. Intercalation, DNA kinking, and the control of transcription. Science 271, 778–784 (1996).
Rost, B., Yachdav, G. & Liu, J. The PredictProtein server. Nucleic Acids Res. 32, W321–W326 (2004).
Riedl, T., Hanaoka, F. & Egly, J.M. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22, 5293–5303 (2003).
Enzlin, J.H. & Schärer, O.D. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 21, 2045–2053 (2002).
McDowell, M.L., Nguyen, T. & Cleaver, J.E. A single-site mutation in the XPAC gene alters photoproduct recognition. Mutagenesis 8, 155–161 (1993).
Kobayashi, T. et al. Mutational analysis of a function of xeroderma pigmentosum group A (XPA) protein in strand specific repair. Nucleic Acids Res. 26, 4662–4668 (1998).
Fujiwara, Y. et al. Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J. Biol. Chem. 274, 20027–20033 (1999).
Janicijevic, A. et al. DNA bending by the human damage recognition complex XPC-HR23B. DNA Repair (Amst.) 2, 325–336 (2003).
Mu, D., Wakasugi, M., Hsu, D.S. & Sancar, A. Characterization of reaction intermediates of human excision repair nuclease. J. Biol. Chem. 272, 28971–28979 (1997).
O'Donovan, A., Davies, A.A., Moggs, J.G., West, S.C. & Wood, R. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature 371, 432–435 (1994).
Ortiz-Lombardia, M. et al. Crystal structure of a DNA Holliday junction. Nat. Struct. Biol. 6, 913–917 (1999).
Isaacs, R.J. & Spielmann, H.P. A model for initial DNA lesion recognition by NER and MMR based on local conformational flexibility. DNA Repair (Amst.) 3, 455–464 (2004).
Weir, H.M. et al. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 12, 1311–1319 (1993).
Read, C.M., Cary, P.D., Crane-Robinson, C., Driscoll, P.C. & Norman, D.G. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 21, 3427–3436 (1993).
Kasparkova, J., Mellish, K.J., Qu, Y. & Brabec, V. Site-specific d(GpG) intrastrand cross-links formed by dinuclear platinum complexes. Bending and NMR studies. Biochemistry 35, 16705–16713 (1996).
Missura, M. et al. Double-check probing of DNA binding and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 20, 3554–3564 (2001).
Dip, R. & Naegeli, H. Binding of the DNA-dependent protein kinase catalytic subunit to Holliday junctions. Biochem. J. 381, 165–174 (2004).
Ford, J.M. & Hanawalt, P.C. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc. Natl. Acad. Sci. USA 92, 8876–8880 (1995).
Acknowledgements
We thank M. Vitanescu for excellent technical assistance, J. Kaspàrkovà (Academy of Sciences of the Czech Republic) for the gift of cisplatin-modified oligonucleotides and O. Schärer (State University of New York, Stony Brook) for the gift of XPF–ERCC1. This work was supported by Swiss National Science Foundation grant 3100A-101747.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Screening of XPA single mutants (PDF 84 kb)
Supplementary Fig. 2
Purity of XPA proteins (PDF 86 kb)
Supplementary Fig. 3
Sequence alignment of XPA homologs (PDF 45 kb)
Supplementary Table 1
Oligonucleotide primers for mutagenesis (PDF 56 kb)
Rights and permissions
About this article
Cite this article
Camenisch, U., Dip, R., Schumacher, S. et al. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat Struct Mol Biol 13, 278–284 (2006). https://doi.org/10.1038/nsmb1061
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1061
This article is cited by
-
A nuclease-mimetic platinum nanozyme induces concurrent DNA platination and oxidative cleavage to overcome cancer drug resistance
Nature Communications (2022)
-
Single molecule analysis reveals monomeric XPA bends DNA and undergoes episodic linear diffusion during damage search
Nature Communications (2020)
-
DNA damage-induced histone H1 ubiquitylation is mediated by HUWE1 and stimulates the RNF8-RNF168 pathway
Scientific Reports (2017)
-
Minnelide Overcomes Oxaliplatin Resistance by Downregulating the DNA Repair Pathway in Pancreatic Cancer
Journal of Gastrointestinal Surgery (2016)
-
SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair
Nature Communications (2015)