Abstract
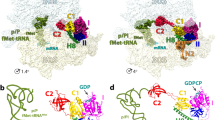
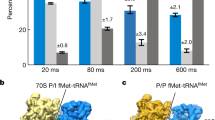
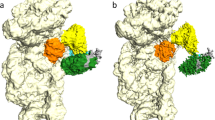
Initiation of protein synthesis is a universally conserved event that requires initiation factors IF1, IF2 and IF3 in prokaryotes. IF2 is a GTPase essential for binding initiator transfer RNA to the 30S ribosomal subunit and recruiting the 50S subunit into the 70S initiation complex. We present two cryo-EM structures of the assembled 70S initiation complex comprising mRNA, fMet-tRNAfMet and IF2 with either a non-hydrolyzable GTP analog or GDP. Transition from the GTP-bound to the GDP-bound state involves substantial conformational changes of IF2 and of the entire ribosome. In the GTP analog–bound state, IF2 interacts mostly with the 30S subunit and extends to the initiator tRNA in the peptidyl (P) site, whereas in the GDP-bound state IF2 steps back and adopts a 'ready-to-leave' conformation. Our data also provide insights into the molecular mechanism guiding release of IF1 and IF3.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Gualerzi, C.O. et al. Initiation factors in the early events of mRNA translation in bacteria. Cold Spring Harb. Symp. Quant. Biol. 66, 363–376 (2001).
Boelens, R. & Gualerzi, C.O. Structure and function of bacterial initiation factors. Curr. Protein Pept. Sci. 3, 107–119 (2002).
Laursen, B.S., Sorensen, H.P., Mortensen, K.K. & Sperling-Petersen, H.U. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69, 101–123 (2005).
Pestova, T.V. et al. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403, 332–335 (2000).
Yusupov, M.M. et al. Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896 (2001).
Jenner, L. et al. Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding. Science 308, 120–123 (2005).
Roll-Mecak, A., Cao, C., Dever, T.E. & Burley, S.K. X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell 103, 781–792 (2000).
Stark, H. et al. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389, 403–406 (1997).
Valle, M. et al. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 21, 3557–3567 (2002).
Stark, H. et al. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 9, 849–854 (2002).
Agrawal, R.K., Penczek, P., Grassucci, R.A. & Frank, J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl. Acad. Sci. USA 95, 6134–6138 (1998).
Stark, H., Rodnina, M.V., Wieden, H.J., van Heel, M. & Wintermeyer, W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell 100, 301–309 (2000).
Klaholz, B.P., Myasnikov, A.G. & van Heel, M. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature 427, 862–865 (2004).
La Teana, A., Gualerzi, C.O. & Dahlberg, A.E. Initiation factor IF 2 binds to the alpha-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA 7, 1173–1179 (2001).
Marzi, S. et al. Ribosomal localization of translation initiation factor IF2. RNA 9, 958–969 (2003).
Diaconu, M. et al. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 121, 991–1004 (2005).
Agrawal, R.K., Heagle, A.B., Penczek, P., Grassucci, R.A. & Frank, J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 6, 643–647 (1999).
Moazed, D., Samaha, R.R., Gualerzi, C. & Noller, H.F. Specific protection of 16S rRNA by translational initiation factors. J. Mol. Biol. 248, 207–210 (1995).
Guenneugues, M. et al. Mapping the fMet-tRNA(f)(Met) binding site of initiation factor IF2. EMBO J. 19, 5233–5240 (2000).
Carter, A.P. et al. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291, 498–501 (2001).
Allen, G.S., Zavialov, A., Gursky, R., Ehrenberg, M. & Frank, J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121, 703–712 (2005).
Martemyanov, K.A., Liljas, A. & Gudkov, A.T. Extremely thermostable elongation factor G from A. aeolicus: cloning, expression, purification, and characterization in a heterologous translation system. Protein Expr. Purif. 18, 257–261 (2000).
Blank, J., Grillenbeck, N.W., Kreutzer, R. & Sprinzl, M. Overexpression and purification of T. thermophilus elongation factors G, Tu, and Ts from E. coli. Protein Expr. Purif. 6, 637–645 (1995).
Rodnina, M.V., Semenkov, Y.P. & Wintermeyer, W. Purification of fMet-tRNA(fMet) by fast protein liquid chromatography. Anal. Biochem. 219, 380–381 (1994).
Gogia, Z.V., Yusupov, M.M. & Spirina, T.N. Structure of Thermus thermophilus ribosomes. 1. Method of isolation and purification of ribosomes. Molekul. Biol. (USSR) 20, 519 (1986).
Yusupova, G.Z., Yusupov, M.M., Cate, J.H. & Noller, H.F. The path of the messenger RNA through the ribosome. Cell 106, 233 (2001).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
van Heel, M. et al. Single-particle cryo electron microscopy: towards atomic resolution. Q. Rev. Biophys. 33, 307–369 (2000).
van Heel, M., Harauz, G., Orlova, E.V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Klaholz, B.P. et al. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature 421, 90–94 (2003).
Rosenthal, P.B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
van Heel, M. & Schatz, M. Fourier shell correlation threshold criteria. J. Struct. Biol. 151, 250–262 (2005).
Plewniak, F. et al. PipeAlign: a new toolkit for protein family analysis. Nucleic Acids Res. 31, 3829–3832 (2003).
Guex, N. & Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997).
Ouali, M. & King, R.D. Cascaded multiple classifiers for secondary structure prediction. Protein Sci. 9, 1162–1176 (2000).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Acknowledgements
We thank J. Thompson (Brown University, Providence, Rhode Island, USA) for providing IF2 strains and D. Moras, P. Schultz, J.-C. Thierry and V. Mallouh for sustained support. We are grateful to L. Jenner for comments on the manuscript and to M. Schatz and R. Schmidt for support with the IMAGIC-5 software. A.G.M. and S.M. are recipients of a postdoctoral fellowship from the CNRS, and A.S. benefited from a fellowship from the Camerino University exchange program. The electron microscope was financed by the Alsace Region, the INSERM, the CNRS and the Association pour la Recherche sur le Cancer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Pair-wise sequence alignment between T. Thermophilus and M thermoautotrophicum IF2/eIF5B sequences. (PDF 228 kb)
Supplementary Fig. 2
Stereo representations of hypothetical models concerning release of IF1 and IF2 during translation initiation. (PDF 791 kb)
Rights and permissions
About this article
Cite this article
Myasnikov, A., Marzi, S., Simonetti, A. et al. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat Struct Mol Biol 12, 1145–1149 (2005). https://doi.org/10.1038/nsmb1012
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1012
This article is cited by
-
Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1
Nature Communications (2018)
-
A conformational switch in initiation factor 2 controls the fidelity of translation initiation in bacteria
Nature Communications (2017)
-
Essential functions linked with structural disorder in organisms of minimal genome
Biology Direct (2016)
-
Initiation of mRNA translation in bacteria: structural and dynamic aspects
Cellular and Molecular Life Sciences (2015)
-
Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA
Nature Structural & Molecular Biology (2014)