Abstract
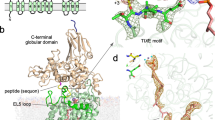
Oligosaccharyltransferase (OST) is a membrane-integral enzyme that catalyzes the transfer of glycans from lipid-linked oligosaccharides (LLOs) onto asparagine side chains, the first step in protein N-glycosylation. Here, we report the X-ray structure of a single-subunit OST, PglB from Campylobacter lari, trapped in an intermediate state bound to an acceptor peptide and a synthetic LLO analog. The structure reveals the role of the external loop EL5, present in all OST enzymes, in substrate recognition. Whereas the N-terminal half of EL5 binds LLO, the C-terminal half interacts with the acceptor peptide. The glycan moiety of LLO must thread under EL5 to access the active site. Reducing EL5 mobility decreases the catalytic rate of OST when full-size heptasaccharide LLO is provided, but not for a monosaccharide-containing LLO analog. Our results define the chemistry of a ternary complex state, assign functional roles to conserved OST motifs, and provide opportunities for glycoengineering by rational design of PglB.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Schwarz, F. & Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582 (2011).
Nothaft, H. & Szymanski, C.M. Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 (2010).
Calo, D., Kaminski, L. & Eichler, J. Protein glycosylation in Archaea: sweet and extreme. Glycobiology 20, 1065–1076 (2010).
Szymanski, C.M., Yao, R., Ewing, C.P., Trust, T.J. & Guerry, P. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32, 1022–1030 (1999).
Helenius, A. & Aebi, M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 (2004).
Kornfeld, R. & Kornfeld, S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54, 631–664 (1985).
Cherepanova, N., Shrimal, S. & Gilmore, R. N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr. Opin. Cell Biol. 41, 57–65 (2016).
Nothaft, H. & Szymanski, C.M. Bacterial protein N-glycosylation: new perspectives and applications. J. Biol. Chem. 288, 6912–6920 (2013).
Valguarnera, E., Kinsella, R.L. & Feldman, M.F. Sugar and spice make bacteria not nice: protein glycosylation and its influence in pathogenesis. J. Mol. Biol. 428, 3206–3220 (2016).
Wacker, M. et al. Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc. Natl. Acad. Sci. USA 103, 7088–7093 (2006).
Marshall, R.D. Glycoproteins. Annu. Rev. Biochem. 41, 673–702 (1972).
Lairson, L.L., Henrissat, B., Davies, G.J. & Withers, S.G. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 (2008).
Liu, J. & Mushegian, A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 12, 1418–1431 (2003).
Kelleher, D.J. & Gilmore, R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R (2006).
Yan, Q. & Lennarz, W.J. Studies on the function of oligosaccharyl transferase subunits: a glycosylatable photoprobe binds to the luminal domain of Ost1p. Proc. Natl. Acad. Sci. USA 99, 15994–15999 (2002).
Nasab, F.P., Schulz, B.L., Gamarro, F., Parodi, A.J. & Aebi, M. All in one: Leishmania major STT3 proteins substitute for the whole oligosaccharyltransferase complex in Saccharomyces cerevisiae. Mol. Biol. Cell 19, 3758–3768 (2008).
Lizak, C., Gerber, S., Numao, S., Aebi, M. & Locher, K.P. X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 (2011).
Matsumoto, S. et al. Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc. Natl. Acad. Sci. USA 110, 17868–17873 (2013).
Matsumoto, S., Taguchi, Y., Shimada, A., Igura, M. & Kohda, D. Tethering an N-glycosylation sequon-containing peptide creates a catalytically competent oligosaccharyltransferase complex. Biochemistry 56, 602–611 (2017).
Gerber, S. et al. Mechanism of bacterial oligosaccharyltransferase: in vitro quantification of sequon binding and catalysis. J. Biol. Chem. 288, 8849–8861 (2013).
Lizak, C. et al. Unexpected reactivity and mechanism of carboxamide activation in bacterial N-linked protein glycosylation. Nat. Commun. 4, 2627 (2013).
Bause, E., Breuer, W. & Peters, S. Investigation of the active site of oligosaccharyltransferase from pig liver using synthetic tripeptides as tools. Biochem. J. 312, 979–985 (1995).
Imperiali, B., Shannon, K.L. & Rickert, K.W. Role of peptide conformation in asparagine-linked glycosylation. J. Am. Chem. Soc. 114, 7942–7944 (1992).
Imperiali, B. & Tai, V.W. in Carbohydrate-Based Drug Discovery (ed. C.-H. Wong) 281–303 (Wiley-VCH, 2003).
Liu, F. et al. Rationally designed short polyisoprenol-linked PglB substrates for engineered polypeptide and protein N-glycosylation. J. Am. Chem. Soc. 136, 566–569 (2014).
Musumeci, M.A. et al. In vitro activity of Neisseria meningitidis PglL O-oligosaccharyltransferase with diverse synthetic lipid donors and a UDP-activated sugar. J. Biol. Chem. 288, 10578–10587 (2013).
Compain, P. & Martin, O.R. Carbohydrate mimetics-based glycosyltransferase inhibitors. Bioorg. Med. Chem. 9, 3077–3092 (2001).
Ramírez, A.S. et al. Characterization of the single-subunit oligosaccharyltransferase STT3A from Trypanosoma brucei using synthetic peptides and lipid-linked oligosaccharide analogs. Glycobiology 27, 525–535 (2017).
Hajduch, J. et al. A convenient synthesis of the C-1-phosphonate analogue of UDP-GlcNAc and its evaluation as an inhibitor of O-linked GlcNAc transferase (OGT). Carbohydr. Res. 343, 189–195 (2008).
Knapp, S. & Myers, D.S. α-GlcNAc thioconjugates. J. Org. Chem. 66, 3636–3638 (2001).
Knapp, S. & Ajayi, K. The anomeric Pudovik rearrangement. Tetrahedr. Lett. 48, 1945–1949 (2007).
Knapp, S., Gonzalez, S., Myers, D.S., Eckman, L.L. & Bewley, C.A. Shortcut to mycothiol analogues. Org. Lett. 4, 4337–4339 (2002).
Engel, R. Phosphonates as analogues of natural phosphates. Chem. Rev. 77, 349–367 (1977).
Jaffee, M.B. & Imperiali, B. Exploiting topological constraints to reveal buried sequence motifs in the membrane-bound N-linked oligosaccharyl transferases. Biochemistry 50, 7557–7567 (2011).
Ihssen, J. et al. Structural insights from random mutagenesis of Campylobacter jejuni oligosaccharyltransferase PglB. BMC Biotechnol. 12, 67 (2012).
Lizak, C. et al. A catalytically essential motif in external loop 5 of the bacterial oligosaccharyltransferase PglB. J. Biol. Chem. 289, 735–746 (2014).
Weerapana, E., Glover, K.J., Chen, M.M. & Imperiali, B. Investigating bacterial N-linked glycosylation: synthesis and glycosyl acceptor activity of the undecaprenyl pyrophosphate-linked bacillosamine. J. Am. Chem. Soc. 127, 13766–13767 (2005).
Glover, K.J., Weerapana, E. & Imperiali, B. In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. USA 102, 14255–14259 (2005).
Igura, M. et al. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 27, 234–243 (2008).
Pedebos, C., Arantes, P.R., Giesel, G.M. & Verli, H. In silico investigation of the PglB active site reveals transient catalytic states and octahedral metal ion coordination. Glycobiology 25, 1183–1195 (2015).
Lee, S.H. & Im, W. Transmembrane motions of PglB induced by LLO are coupled with EL5 loop conformational changes necessary for OST activity. Glycobiology 27, 734–742 (2017).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 (2002).
DeLano, W.L. The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
Kowarik, M. et al. N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science 314, 1148–1150 (2006).
Perez, C. et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433–438 (2015).
Acknowledgements
This research was supported by the Swiss National Science Foundation (SNF 310030B_166672 to K.P.L. and Transglyco Sinergia grant to M.A., J.-L.R. and K.P.L.). We thank the beamline staff at the Swiss Light Source for assistance with data collection, C. Lizak for preparing a cysteineless construct of PglB, and A. Ramírez for assistance with the chemo-enzymatic synthesis of farnesyl-PP-GlcNAc-1,3α-GalNAc.
Author information
Authors and Affiliations
Contributions
M.N. performed the overexpression, purification, disulfide cross-linking, functional characterization, and crystallization of PglB. K.P.L. and M.N. performed X-ray data collection, structure determination, and model building and refinement. J.B., T.S., T.D., and J.-L.R. synthesized LLO analogs. M.N., M.A., and K.P.L. devised experiments and analyzed the data. K.P.L. and M.N. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
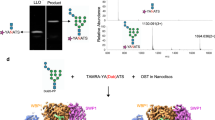
Supplementary Figure 1 In vitro activity of PglB with synthetic LLO analogs.
PglB activity was measured in vitro using synthetic LLO analogs with distinct polyprenyl tails. (a) Structures of synthetic LLO analogs featuring cis (ωZ-PP-GlcNAc) or trans (ωE-PP-GlcNAc) configuration of double bonds. (b) Tricine SDS-PAGE analysis of peptide glycosylation determined by quantification of fluorescently labeled substrate. The assays were performed at 30 °C and contained 1 μM PglB and 100 μM synthetic LLO analog. (c) Turnover numbers of glycosylation assays shown in b. Data represent three independent protein preparations (error bars indicate s.d., n=3).
Supplementary Figure 2 Structures of lipid-linked oligosaccharides.
(a) Structure of full-length, wild type C. jejuni LLO, abbreviated as undecaprenyl-PP-BacGalNAc5Glc. (b) Structure of synthetic, inhibitory LLO nerylneryl-PPC-GlcNAc as used for co-crystallization and structure determiation of PglB ternary complex.
Supplementary Figure 3 Kinetic analysis of synthetic LLO substrates.
Normalized PglB activities as a function of varying concentrations of three LLO analogs (structures shown in Fig. 1a). Activities were normalized to the maximal turnover rate for each LLO analog independently to allow the comparison of the Michaelis constants KM. Each data point represents three independent cell cultures, calculated from the slope of the linear regression fit (error bars indicate s.d., n≥4)
Supplementary Figure 4 PglB membrane topology.
Topological scheme of PglB transmembrane domain, with TM helices depicted as cylinders and numbered. Dashed green lines indicate non-covalent contacts to the periplasmic domain. The N-terminal and C-terminal segments of the external loop EL5 are indicated as N-EL5 and C-EL5, respectively.
Supplementary Figure 5 Surface representation of the ternary complex structure. PglB- acceptor peptide-LLO complex.
(a) Surface representation of the ternary complex structure of PglB bound to acceptor peptide and synthetic, inhibitory LLO analog. PglB is colored grey, N-EL5 in turquoise, C-EL5 in purple, bound peptide in orange, bound LLO in yellow. (b) Close-up view of a. The acceptor peptide and inhibitory LLO are shown in orange and yellow sticks, respectively. The substrates are bound to cavities at opposite entrances / exits of the PglB tunnel.
Supplementary Figure 6 Catalytic site and acceptor asparagine binding.
(a) Stereo view of the catalytic site and acceptor peptide binding. PglB is colored as in Fig. 2a, selected residues are shown as sticks and labeled. The acceptor peptide is shown as orange sticks, with residues numbered relative to their position in the acceptor sequon (Asn at position zero). The bound Mn2+ ion is shown as a pink sphere and labeled. (b) Schematic view of coordination geometry of Mn2+ ion, yellow dashed lines depict octahedral shape. The six ligands are the side-chain atoms of D56, D154, E319 and three water molecules, shown as red spheres.
Supplementary Figure 7 Disulfide cross-linking efficiency of the cysteine double mutant.
The ratio of disulfide cross-linking was determined by labeling unreacted, free cysteines with fluorescein maleimide and quantitation of in-gel fluorescence. (a) Coomassie-stained gel; (b) fluorescence-scanned gel. The oxidative disulfide bond formation was determined to be 80 +/- 2.9% from three independent cell cultures (error denotes s.d., n=3). Lanes labeled “ox” (oxidized) indicates a sample that was cross-linked with CuCl2 during the purification process, whereas “red” (reduced) indicates a control sample that contained 10 mM β-mercaptoethanol during purification, which was removed by desalting. MW denotes marker proteins, with masses indicated on the side.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Note 1 (PDF 1049 kb)
Supplementary Data Set 1
Uncropped gel image for Fig 4b (PDF 133 kb)
Rights and permissions
About this article
Cite this article
Napiórkowska, M., Boilevin, J., Sovdat, T. et al. Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase. Nat Struct Mol Biol 24, 1100–1106 (2017). https://doi.org/10.1038/nsmb.3491
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3491
This article is cited by
-
Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase
Nature Chemical Biology (2023)
-
Structure of the human heparan sulfate polymerase complex EXT1-EXT2
Nature Communications (2022)
-
Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase
Nature Communications (2022)
-
Structural basis of lipopolysaccharide maturation by the O-antigen ligase
Nature (2022)
-
Conserved sequence motifs in human TMTC1, TMTC2, TMTC3, and TMTC4, new O-mannosyltransferases from the GT-C/PMT clan, are rationalized as ligand binding sites
Biology Direct (2021)