Abstract
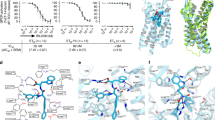
Endothelin receptors (ETRs) have crucial roles in vascular control and are targets for drugs designed to treat circulatory-system diseases and cancer progression. The nonpeptide dual-ETR antagonist bosentan is the first oral drug approved to treat pulmonary arterial hypertension. Here we report crystal structures of human endothelin ETB receptor bound to bosentan and to the ETB-selective analog K-8794, at 3.6-Å and 2.2-Å resolution, respectively. The K-8794-bound structure reveals the detailed water-mediated hydrogen-bonding network at the transmembrane core, which could account for the weak negative allosteric modulation of ETB by Na+ ions. The bosentan-bound structure reveals detailed interactions with ETB, which are probably conserved in the ETA receptor. A comparison of the two structures shows unexpected similarity between antagonist and agonist binding. Despite this similarity, bosentan sterically prevents the inward movement of transmembrane helix 6 (TM6), and thus exerts its antagonistic activity. These structural insights will facilitate the rational design of new ETR-targeting drugs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Yanagisawa, M. et al. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature 332, 411–415 (1988).
Arai, H., Hori, S., Aramori, I., Ohkubo, H. & Nakanishi, S. Cloning and expression of a cDNA encoding an endothelin receptor. Nature 348, 730–732 (1990).
Sakurai, T. et al. Cloning of a cDNA encoding a non-isopeptide-selective subtype of the endothelin receptor. Nature 348, 732–735 (1990).
Kedzierski, R.M. & Yanagisawa, M. Endothelin system: the double-edged sword in health and disease. Annu. Rev. Pharmacol. Toxicol. 41, 851–876 (2001).
Kohan, D.E., Rossi, N.F., Inscho, E.W. & Pollock, D.M. Regulation of blood pressure and salt homeostasis by endothelin. Physiol. Rev. 91, 1–77 (2011).
Rubanyi, G.M. & Polokoff, M.A. Endothelins: molecular biology, biochemistry, pharmacology, physiology, and pathophysiology. Pharmacol. Rev. 46, 325–415 (1994).
Shin, M.K., Levorse, J.M., Ingram, R.S. & Tilghman, S.M. The temporal requirement for endothelin receptor-B signalling during neural crest development. Nature 402, 496–501 (1999).
Remuzzi, G., Perico, N. & Benigni, A. New therapeutics that antagonize endothelin: promises and frustrations. Nat. Rev. Drug Discov. 1, 986–1001 (2002).
Channick, R.N. et al. Effects of the dual endothelin-receptor antagonist bosentan in patients with pulmonary hypertension: a randomised placebo-controlled study. Lancet 358, 1119–1123 (2001).
Rubin, L.J. et al. Bosentan therapy for pulmonary arterial hypertension. N. Engl. J. Med. 346, 896–903 (2002).
Dhaun, N., Goddard, J. & Webb, D.J. The endothelin system and its antagonism in chronic kidney disease. J. Am. Soc. Nephrol. 17, 943–955 (2006).
Rosanò, L., Spinella, F. & Bagnato, A. Endothelin 1 in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer 13, 637–651 (2013).
Clozel, M. et al. Pharmacological characterization of bosentan, a new potent orally active nonpeptide endothelin receptor antagonist. J. Pharmacol. Exp. Ther. 270, 228–235 (1994).
Clozel, M. et al. Pathophysiological role of endothelin revealed by the first orally active endothelin receptor antagonist. Nature 365, 759–761 (1993).
Neidhart, W. et al. The discovery of nonpeptide endothelin receptor antagonists. Progression towards bosentan. Chimia 50, 519–524 (1996).
Mucke, H.A. Pulmonary arterial hypertension: on the way to a manageable disease. Curr. Opin. Investig. Drugs 9, 957–962 (2008).
Norman, P. Pulmonary arterial hypertension: a rare disease that encourages the development of multiple treatments. Expert Opin. Orphan Drugs 2, 1137–1145 (2014).
Korn, J.H. et al. Digital ulcers in systemic sclerosis: prevention by treatment with bosentan, an oral endothelin receptor antagonist. Arthritis Rheum. 50, 3985–3993 (2004).
Gatfield, J., Mueller Grandjean, C., Bur, D., Bolli, M.H. & Nayler, O. Distinct ETA receptor binding mode of macitentan as determined by site directed mutagenesis. PLoS One 9, e107809 (2014).
Gatfield, J., Mueller Grandjean, C., Sasse, T., Clozel, M. & Nayler, O. Slow receptor dissociation kinetics differentiate macitentan from other endothelin receptor antagonists in pulmonary arterial smooth muscle cells. PLoS One 7, e47662 (2012).
Krum, H., Viskoper, R.J., Lacourciere, Y., Budde, M. & Charlon, V. The effect of an endothelin-receptor antagonist, bosentan, on blood pressure in patients with essential hypertension. N. Engl. J. Med. 338, 784–790 (1998).
Bolli, M.H. et al. The discovery of N-[5-(4-bromophenyl)-6-[2-[(5-bromo-2-pyrimidinyl)oxy]ethoxy]-4-pyrimidinyl]-N′-propylsulfamide (macitentan), an orally active, potent dual endothelin receptor antagonist. J. Med. Chem. 55, 7849–7861 (2012).
Vatter, H. & Seifert, V. Ambrisentan, a non-peptide endothelin receptor antagonist. Cardiovasc. Drug Rev. 24, 63–76 (2006).
Barst, R.J. et al. Sitaxsentan therapy for pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 169, 441–447 (2004).
Boss, C., Bolli, M.H. & Gatfield, J. From bosentan (Tracleer®) to macitentan (Opsumit®): the medicinal chemistry perspective. Bioorg. Med. Chem. Lett. 26, 3381–3394 (2016).
Kholdani, C.A., Fares, W.H. & Trow, T.K. Macitentan for the treatment of pulmonary arterial hypertension. Vasc. Health Risk Manag. 10, 665–673 (2014).
Shihoya, W. et al. Activation mechanism of endothelin ETB receptor by endothelin-1. Nature 537, 363–368 (2016).
Sawaki, M. et al. Chronic effects of an orally active selective endothelin-B-receptor antagonist in experimental congestive heart failure. J. Cardiovasc. Pharmacol. 36, S323–S326 (2000).
Okuta, A., Tani, K., Nishimura, S., Fujiyoshi, Y. & Doi, T. Thermostabilization of the human endothelin type B receptor. J. Mol. Biol. 428, 2265–2274 (2016).
Hattori, M., Hibbs, R.E. & Gouaux, E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure 20, 1293–1299 (2012).
Parker, M.S., Wong, Y.Y. & Parker, S.L. An ion-responsive motif in the second transmembrane segment of rhodopsin-like receptors. Amino Acids 35, 1–15 (2008).
Katritch, V. et al. Allosteric sodium in class A GPCR signaling. Trends Biochem. Sci. 39, 233–244 (2014).
Liu, W. et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337, 232–236 (2012).
Miller-Gallacher, J.L. et al. The 2.1 Å resolution structure of cyanopindolol-bound β1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor. PLoS One 9, e92727 (2014).
Fenalti, G. et al. Molecular control of δ-opioid receptor signalling. Nature 506, 191–196 (2014).
Zhang, C. et al. High-resolution crystal structure of human protease-activated receptor 1. Nature 492, 387–392 (2012).
Harding, M.M. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr. D Biol. Crystallogr. 58, 872–874 (2002).
White, J.F. et al. Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513 (2012).
Yin, J., Mobarec, J.C., Kolb, P. & Rosenbaum, D.M. Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature 519, 247–250 (2015).
Thompson, A.A. et al. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature 485, 395–399 (2012).
Takasuka, T., Sakurai, T., Goto, K., Furuichi, Y. & Watanabe, T. Human endothelin receptor ETB. Amino acid sequence requirements for super stable complex formation with its ligand. J. Biol. Chem. 269, 7509–7513 (1994).
Rose, P.M. et al. Aspartate mutation distinguishes ETA but not ETB receptor subtype-selective ligand binding while abolishing phospholipase C activation in both receptors. FEBS Lett. 361, 243–249 (1995).
Opgenorth, T.J. et al. Pharmacological characterization of A-127722: an orally active and highly potent ETA-selective receptor antagonist. J. Pharmacol. Exp. Ther. 276, 473–481 (1996).
Winn, M. et al. 2,4-Diarylpyrrolidine-3-carboxylic acids—potent ETA selective endothelin receptor antagonists. 1. Discovery of A-127722. J. Med. Chem. 39, 1039–1048 (1996).
Kikuchi, T. et al. Endothelin-1 analogues substituted at both position 18 and 19: highly potent endothelin antagonists with no selectivity for either receptor subtype ETA or ETB . J. Med. Chem. 36, 4087–4093 (1993).
Tam, J.P. et al. Alanine scan of endothelin: importance of aromatic residues. Peptides 15, 703–708 (1994).
Rovero, P., Patacchini, R. & Maggi, C.A. Structure-activity studies on endothelin (16-21), the C-terminal hexapeptide of the endothelins, in the guinea-pig bronchus. Br. J. Pharmacol. 101, 232–234 (1990).
Henry, J.A., Horwell, D.C., Meecham, K.G. & Rees, D.C. A structure-affinity study of the amino-acid side-chains in neurotensin—N and C-terminal deletions and Ala-scan. Bioorg. Med. Chem. Lett. 3, 949–952 (1993).
Guerrini, R. et al. Address and message sequences for the nociceptin receptor: a structure-activity study of nociceptin-(1-13)-peptide amide. J. Med. Chem. 40, 1789–1793 (1997).
Nakajima, K. et al. Structure-activity relationship of endothelin: importance of charged groups. Biochem. Biophys. Res. Commun. 163, 424–429 (1989).
Vagner, J., Qu, H. & Hruby, V.J. Peptidomimetics, a synthetic tool of drug discovery. Curr. Opin. Chem. Biol. 12, 292–296 (2008).
Hruby, V.J. Designing peptide receptor agonists and antagonists. Nat. Rev. Drug Discov. 1, 847–858 (2002).
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Ueno, G. et al. Remote access and automation of SPring-8 MX beamlines. AIP Conf. Proc. 1741, 050021 (2016).
Kabsch, W. Processing of X-ray snapshots from crystals in random orientations. Acta Crystallogr. D Biol. Crystallogr. 70, 2204–2216 (2014).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Inoue, A. et al. TGFα shedding assay: an accurate and versatile method for detecting GPCR activation. Nat. Methods 9, 1021–1029 (2012).
Doi, T., Sugimoto, H., Arimoto, I., Hiroaki, Y. & Fujiyoshi, Y. Interactions of endothelin receptor subtypes A and B with Gi, Go, and Gq in reconstituted phospholipid vesicles. Biochemistry 38, 3090–3099 (1999).
Wada, K. et al. Purification of an endothelin receptor from human placenta. Biochem. Biophys. Res. Commun. 167, 251–257 (1990).
Elshourbagy, N.A. et al. Molecular cloning and characterization of the major endothelin receptor subtype in porcine cerebellum. Mol. Pharmacol. 41, 465–473 (1992).
Acknowledgements
We thank the members of the Nureki lab and the beamline staff at BL32XU of SPring-8 (Sayo, Japan) for technical assistance during data collection. We also thank Kowa Co., Ltd., for providing K-8794. pCAGGS expression plasmid vector was a kind gift from J. Miyazaki (Osaka University, Osaka, Japan). The diffraction experiments were performed at SPring-8 BL32XU (proposals 2015A1024, 2015A1057, 2015B2024, and 2015B2057). This work was supported by JSPS KAKENHI grants 16K07172 (T.D.), 26640102 (T.D.), 16H06294 (O.N.), 15H05775 (F.Y.), 15J09780 (S.W.), 17J30010 (S.W.), 17H05000 (T.N.) and 15H06862 (K.Y.), the Core Research for Evolutional Science, PRESTO from the Japan Science and Technology (JST) Technology Program; the Platform for Drug Discovery, Information, and Structural Life Science from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Japan Agency for Medical Research and Development (AMED); and the National Institute of Biomedical Innovation. A.I. was funded by JST, PRESTO (grant JPMJPR1331), and the PRIME from AMED. J.A. received funding from AMED-CREST and AMED, and a MEXT Grant-in-Aid for Scientific Research on Innovative Areas (grant 15H05897).
Author information
Authors and Affiliations
Contributions
W.S. designed experiments; expressed, purified, and crystallized the antagonist-bound ETB receptor; collected data; and refined the structures. T.N. initially crystallized the K-8794-bound ETB receptor, assisted with the structural determination, and designed the construct ETB-Y4-mT4L. K.Y. and K.H. developed a pipeline for data collection and processing, and assisted with the structural determination. A.I., F.M.N.K., and J.A. performed and oversaw the cell-based assays. A.O. introduced K-8794 in the experimental design and characterized its pharmacology. K.T. initially designed the T4L-fused construct. T.D. performed the radiobinding assays. The manuscript was prepared by W.S., T.N., K.Y., A.I., K.H., K.T., Y.F., T.D., and O.N. Y.F., T.D., and O.N. supervised the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Crystallization.
a, Crystallization constructs of the ETB receptor are shown, with all of the modifications to the human wild-type the ETB receptor indicated. Residues interacting with both bosentan and K-8794 are colored green, the residue only interacting with bosentan is blue, and those only interacting with K-8794 are red, as shown in the figure. b, c, Effects of ET-1 on the release of AP-TGFα and antagonists on the ET-1-induced release of AP-TGFα in HEK293 cells expressing the endothelin receptors. In the competitive assays, the concentration of the agonist ET-1 was 0.2 nM, and the AP-TGFα release response in the ET-1 treatment alone was normalized to 100%. Symbols and error bars are means and s.e.m. (standard error of the mean), respectively. For most data points, the error bars are smaller than the symbols. d, e, Crystallographic data of the ETB-Y5-mT4L protein bound to K-8794 (d) and the ETB-Y4-mT4L protein bound to bosentan (e). The left panels show the crystals of the antagonist-bound ETB receptors. The middle and right panels show their crystal packings. T4L is shown as a grey cartoon, and the K-8794- and bosentan-bound ETB receptors are shown as orange and turquoise cartoons, respectively. Crystal lattices are indicated by black lines.
Supplementary Figure 2 Electron density.
a, b, Fo − Fc omit maps for K-8794 (a) and bosentan (b), contoured at 3.0 σ and 4.0 σ, respectively. TM6 and TM7 are omitted. c, The bosentan binding site, in which the colors represent the temperature factors ranging from 20 Å2 (blue) to 120 Å2 (red). d, Stereo view of the 2Fo−Fc map, contoured at 1.0 σ, for the residues within 4 Å contact distances of the ligand in the K-8794-bound ETB structure. e, Stereo views of the 2Fo−Fc maps, contoured at 1.0 σ, for the residues within 4 Å contact distances of the ligand in the bosentan-bound ETB structure. f, Stereo view of the composite omit map, contoured at 1.0 σ, for the residues within 4 Å contact distances of the ligand in the bosentan-bound ETB structure.
Supplementary Figure 3 Comparison with other peptide-activated GPCRs.
a–c, Comparison of the antagonist binding sites of the peptide-activated GPCRs. Ribbon representations of the ETB receptor in complex with bosentan (a), Orexin receptor OX2 in complex with suvorexant (PDB accession number 4RNB) (b), and NOP receptor in complex with the peptidomimetic antagonist C-24 (PDB accession number 4EA3) (c) are aligned according to the position of Trp6.48, which is indicated by the stick model in each figure. The black dashed line indicates the position of the Cα atoms of Trp6.48. The small-molecule antagonists are represented by stick models. Like the ETB receptor, OX2 belongs to the β subfamily of the class A GPCRs, while NOP belongs to the γ subfamily. d, e, Electrostatic surfaces of the ETB structures bound to bosentan (d) and ET-1 (e), viewed from the extracellular side (left) and within the membrane plane (right). Bosentan and ET-1 are shown as sticks and transparent surfaces, colored blue and pink, respectively.
Supplementary Figure 4 Ligand-interaction diagrams.
Interaction diagrams of K-8794 (a) and bosentan (b) with the ETB receptor. Interactions within 4 Å are shown. Polar and hydrophobic contacts are represented as red dashed and green lines, respectively.
Supplementary Figure 5 Homology between ETB and ETA.
Amino acid sequence alignment of the human ETB (UniProt ID: P24530) and ETA (P25101) receptors. Secondary structure elements for α-helices and β-strands are indicated by cylinders and arrows, respectively. Conservation of the residues between ETA and ETB is indicated as follows: red panels for completely conserved, red letters for partially conserved, and black letters for not conserved. The residues involved in bosentan and K-8794 binding are shown as blue squares and orange diamonds, respectively.
Supplementary Figure 6 Small-molecule endothelin-receptor antagonists.
Chemical structures of major small-molecule endothelin receptor antagonists. Endothelin receptor antagonists commonly have negatively-charged moieties (sulfonamide or carboxylate).
Supplementary Figure 7 Comparison of structural changes on ligand binding.
a, b, Comparison of the ET-1 and bosentan binding modes, coloured as in Fig. 6. Receptors and ET-1 are represented by ribbons, and the side chains of ET-118-21 and bosentan are shown as sticks with transparent surfaces. c, The residues that interact with both ET-1 and bosentan are superimposed. d–f, Comparison of the structural changes upon ET-1 (d), bosentan (e), and K-8794 (f) binding, coloured as in Figs. 1 and 4.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Note 1 (PDF 1765 kb)
Rights and permissions
About this article
Cite this article
Shihoya, W., Nishizawa, T., Yamashita, K. et al. X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog. Nat Struct Mol Biol 24, 758–764 (2017). https://doi.org/10.1038/nsmb.3450
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3450
This article is cited by
-
Structural basis of peptide recognition and activation of endothelin receptors
Nature Communications (2023)
-
Fly casting with ligand sliding and orientational selection supporting complex formation of a GPCR and a middle sized flexible molecule
Scientific Reports (2022)
-
Computer simulation of molecular recognition in biomolecular system: from in silico screening to generalized ensembles
Biophysical Reviews (2022)
-
Cryo-EM structure of the human PAC1 receptor coupled to an engineered heterotrimeric G protein
Nature Structural & Molecular Biology (2020)
-
Crystal structure of human endothelin ETB receptor in complex with peptide inverse agonist IRL2500
Communications Biology (2019)