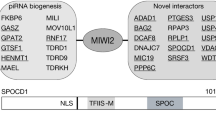
Abstract
In mice, the pathway involving PIWI and PIWI-interacting RNA (PIWI–piRNA) is essential to re-establish transposon silencing during male-germline reprogramming. The cytoplasmic PIWI protein MILI mediates piRNA-guided transposon RNA cleavage as well as piRNA amplification. MIWI2's binding to piRNA and its nuclear localization are proposed to be dependent upon MILI function. Here, we demonstrate the existence of a piRNA biogenesis pathway that sustains partial MIWI2 function and reprogramming activity in the absence of MILI.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Aravin, A.A. et al. Mol. Cell 31, 785–799 (2008).
De Fazio, S. et al. Nature 480, 259–263 (2011).
Carmell, M.A. et al. Dev. Cell 12, 503–514 (2007).
Kuramochi-Miyagawa, S. et al. Genes Dev. 22, 908–917 (2008).
Aravin, A.A., Sachidanandam, R., Girard, A., Fejes-Toth, K. & Hannon, G.J. Science 316, 744–747 (2007).
Kuramochi-Miyagawa, S. et al. Development 131, 839–849 (2004).
Molaro, A. et al. Genes Dev. 28, 1544–1549 (2014).
Manakov, S.A. et al. Cell Rep. 12, 1234–1243 (2015).
Han, B.W., Wang, W., Li, C., Weng, Z. & Zamore, P.D. Science 348, 817–821 (2015).
Mohn, F., Handler, D. & Brennecke, J. Science 348, 812–817 (2015).
Homolka, D. et al. Cell Rep. 12, 418–428 (2015).
Yang, Z. et al. Mol. Cell 61, 138–152 (2016).
Wang, W. et al. Mol. Cell 59, 819–830 (2015).
Saxe, J.P., Chen, M., Zhao, H. & Lin, H. EMBO J. 32, 1869–1885 (2013).
Di Giacomo, M. et al. Mol. Cell 50, 601–608 (2013).
Carrieri, C. et al. J. Exp. Med. http://dx.doi.org/10.1084/jem.20161371 (2017).
Yang, H. et al. Cell 154, 1370–1379 (2013).
Miura, F. & Ito, T. DNA Res. 22, 13–18 (2015).
Krueger, F. & Andrews, S.R. Bioinformatics 27, 1571–1572 (2011).
Nawrocki, E.P. et al. Nucleic Acids Res. 43, D130–D137 (2015).
Davis, M.P., van Dongen, S., Abreu-Goodger, C., Bartonicek, N. & Enright, A.J. Methods 63, 41–49 (2013).
Clèries, R. et al. Comput. Biol. Med. 42, 438–445 (2012).
Acknowledgements
The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. GA 310206 awarded to D.O'C.
Author information
Authors and Affiliations
Contributions
L.V. contributed to the design, execution and analysis of the majority of experiments. D.V. performed the bioinformatic piRNA analysis. R.V.B. generated the whole-genome bisulfite libraries and performed the bioinformatic analysis of genomic methylation. C.C. designed, generated and validated the Miwi2HA allele. A.J.E. and W.R. oversaw all bioinformatic analyses performed. D.O'C. conceived and supervised this study. L.V. and D.O'C. wrote the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 An overview of piRNA biogenesis during fetal mouse reprogramming.
(a) A schematic overview of the production and amplification of a single piRNA from a transposon transcript. The transposon mRNA is processed by the primary piRNA biogenesis machinery to generate primary sense piRNAs that have a 1U bias. Indicated in yellow is a representative piRNA that will be processed from this transcript. Once bound to MILI, the piRNA guides endonucleolytic cleavage of complementary transcripts between 10th and 11th nucleotide counting from a 5′ end of bound piRNA. The 3′ cleavage product will be processed to form a secondary antisense piRNA, the location of which is indicated in blue. The nascent secondary piRNA is loaded into MILI or MIWI2. This licences MIWI2 to enter the nucleus, where it is proposed to tether to a nascent transposon transcript and direct de novo DNA methylation by an unknown mechanism. The binding of the secondary antisense piRNA to MILI results in the piRNA-guided cleavage of transposon transcript and the biogenesis of the original piRNA. This MILI:MILI homotypic ping-pong cleavage cycle amplifies transposon-derived piRNAs during fetal reprogramming. (b) Features of amplified piRNA pairs. Due to the fact that most primary piRNAs exhibit 1U bias, newly generated secondary piRNAs will have a bias for A at their 10th position (10A). Complementarity between the first 10 nucleotides of amplified piRNA pairs is a key signature of the ping-pong cycle.
Supplementary Figure 2 Loss of MILI and MIWI2 has different impact on genomic and repeat element methylation.
Distribution of CpG methylation levels measured by whole-genome bisulfite sequencing of Mili−/− (dark blue) and Miwi2−/− (light blue) spermatogonia plotted as a difference to wild-type (WT) spermatogonia (grey dotted line). Significance was tested using Wilcoxon rank sum test with Bonferroni multiple testing correction, *** indicates a p value of < 0.0005. Graphs show averages of values obtained from three animals. The middle line in the graphs represents median of the data points with whiskers indicating the data range.
Supplementary Figure 3 Transposon-related piRNAs in the absence of MILI.
(a) Length distribution of small RNAs in wild-type (WT) and Mili−/− E16.5 fetal testis. (b) Quantification of LINE1 and IAP associated piRNAs in WT and Mili−/− E16.5 fetal testis. (c) piRNAs from WT and Mili−/− E16.5 fetal testis mapped to LINE1 and IAP consensus sequence allowing three mismatches. Positive (red) and negative (blue) values indicate sense and antisense piRNAs, respectively. (d) Ping-pong analysis of LINE1 and IAP associated piRNAs in WT and Mili−/− E16.5 fetal testis. The relative frequency of the distance between 5′ ends of complementary piRNAs for LINE1 (right) and IAP (left) is presented. (e) Percentage of LINE1 and IAP associated piRNAs in WT and Mili−/− E16.5 fetal testis with a U at the first position (1U) without an A at the position 10 and an A at position 10 (10A) without a U at position 1 is shown. (f) piRNAs from WT and Mili−/− E16.5 fetal testis mapped to loci whose methylation is dependent upon MIWI2 and MILI, respectively. Positive (red) and negative (blue) values indicate sense and antisense piRNAs, respectively. (g) Quantification of all or uniquely mapping piRNA densities across MILI and MIWI2-dependent loci is presented. Significance in e and g was assessed using BootstRatio algorithm, * indicates a p value of < 0.05, ** indicates a p value of < 0.005, *** indicates a p value of < 0.0005. Graphs in a, c, d, f show averages of values obtained from two animals. Individual data points in b, e, g show values obtained from two animals.
Supplementary Figure 4 MIWI2 retains partial nuclear localization in the absence of MILI.
(a & c) Immunofluorescence with anti-MIWI2 antibody (a) and anti-HA antibody (c) on fetal testis of indicated genotypes with a seminiferous tubule shown. (b & d) Quantification of anti-MIWI2 (b) and anti-HA (d) fluorescence signal in the nucleus of indicated genotypes. 150 gonocytes from three animals, 100 and 97 gonocytes from two animals were analyzed in panel b from E16.5 Miwi2−/−, Miwi2+/− and Mili−/− fetal testes, respectively. 104, 104 and 103 gonocytes from two animals were analyzed in panel d from E16.5 Mili+/−, Mili+/−; Miwi2HA/+ and Mili−/−; Miwi2HA/+ fetal testes, respectively. Graphs show mean and s.d.
Supplementary Figure 5 Generation and validation of the Miwi2HA allele.
(a) A targeting strategy for the generation of Miwi2HA allele by CRISPR-Cas9 gene editing technology is presented. (b) Chromatogram showing HA-tag and BamHI sequence insertion at the site of starting ATG codon in Miwi2 locus. (c) Validation strategy of Miwi2HA allele by PCR followed by BamHI restriction is shown. Forward (Fw) and reverse (Rv) primers flanking starting ATG codon amplify 207 bp fragment in presence of Miwi2+ allele and 240 bp fragment in presence of Miwi2HA allele. When 240 bp fragment is digested with BamHI, 87 bp and 153 bp size fragments are generated (upper panel). PCR amplicon digestion with BamHI is shown as described above (lower panel). (d) Normal testicular weight of adult Miwi2HA/+ and Miwi2HA/HA mice is shown. Graph shows mean and s.d. Individual data points are presented.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 943 kb)
Rights and permissions
About this article
Cite this article
Vasiliauskaitė, L., Vitsios, D., Berrens, R. et al. A MILI-independent piRNA biogenesis pathway empowers partial germline reprogramming. Nat Struct Mol Biol 24, 604–606 (2017). https://doi.org/10.1038/nsmb.3413
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3413
This article is cited by
-
SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation
Nature (2020)
-
TEX15 is an essential executor of MIWI2-directed transposon DNA methylation and silencing
Nature Communications (2020)
-
PIWI-interacting RNAs: small RNAs with big functions
Nature Reviews Genetics (2019)
-
Defective germline reprogramming rewires the spermatogonial transcriptome
Nature Structural & Molecular Biology (2018)
-
The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity
Nature Communications (2017)