Abstract
Long interspersed nuclear elements (LINEs) are ubiquitous transposable elements in higher eukaryotes that have a significant role in shaping genomes, owing to their abundance. Here we report that guanine-rich sequences in the 3′ untranslated regions (UTRs) of hominoid-specific LINE-1 elements are coupled with retrotransposon speciation and contribute to retrotransposition through the formation of G-quadruplex (G4) structures. We demonstrate that stabilization of the G4 motif of a human-specific LINE-1 element by small-molecule ligands stimulates retrotransposition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Cordaux, R. & Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10, 691–703 (2009).
Kazazian, H.H. Jr. Mobile DNA: Finding Treasure in Junk (Pearson Education, 2011).
Levin, H.L. & Moran, J.V. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 12, 615–627 (2011).
Erwin, J.A., Marchetto, M.C. & Gage, F.H. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat. Rev. Neurosci. 15, 497–506 (2014).
Ohshima, K. RNA-mediated gene duplication and retroposons: retrogenes, LINEs, SINEs, and sequence specificity. Int. J. Evol. Biol. 2013, 424726 (2013).
Kajikawa, M. & Okada, N. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell 111, 433–444 (2002).
Takahashi, H. & Fujiwara, H. Transplantation of target site specificity by swapping the endonuclease domains of two LINEs. EMBO J. 21, 408–417 (2002).
Hayashi, Y., Kajikawa, M., Matsumoto, T. & Okada, N. Mechanism by which a LINE protein recognizes its 3′ tail RNA. Nucleic Acids Res. 42, 10605–10617 (2014).
Moran, J.V. et al. High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927 (1996).
Howell, R. & Usdin, K. The ability to form intrastrand tetraplexes is an evolutionarily conserved feature of the 3′ end of L1 retrotransposons. Mol. Biol. Evol. 14, 144–155 (1997).
Lexa, M. et al. Guanine quadruplexes are formed by specific regions of human transposable elements. BMC Genomics 15, 1032 (2014).
Bochman, M.L., Paeschke, K. & Zakian, V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780 (2012).
Kejnovsky, E. & Lexa, M. Quadruplex-forming DNA sequences spread by retrotransposons may serve as genome regulators. Mob. Genet. Elements 4, e28084 (2014).
Kejnovsky, E., Tokan, V. & Lexa, M. Transposable elements and G-quadruplexes. Chromosome Res. 23, 615–623 (2015).
Huppert, J.L. & Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916 (2005).
Chambers, V.S. et al. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 33, 877–881 (2015).
Boissinot, S., Davis, J., Entezam, A., Petrov, D. & Furano, A.V. Fitness cost of LINE-1 (L1) activity in humans. Proc. Natl. Acad. Sci. USA 103, 9590–9594 (2006).
Ostertag, E.M., Prak, E.T., DeBerardinis, R.J., Moran, J.V. & Kazazian, H.H. Jr. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28, 1418–1423 (2000).
Farkash, E.A., Kao, G.D., Horman, S.R. & Prak, E.T. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 34, 1196–1204 (2006).
Rodriguez, R. et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 8, 301–310 (2012).
Piazza, A. et al. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 38, 4337–4348 (2010).
Riou, J.F. et al. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl. Acad. Sci. USA 99, 2672–2677 (2002).
Kale, S.P., Moore, L., Deininger, P.L. & Roy-Engel, A.M. Heavy metals stimulate human LINE-1 retrotransposition. Int. J. Environ. Res. Public Health 2, 14–23 (2005).
Ostertag, E.M. & Kazazian, H.H. Jr. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35, 501–538 (2001).
Khan, H., Smit, A. & Boissinot, S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 16, 78–87 (2006).
Métifiot, M., Amrane, S., Litvak, S. & Andreola, M.L. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res. 42, 12352–12366 (2014).
Smit, A.F.A., Hubley, R. & Green, P. RepeatMasker Open-4.0 (Institute for Systems Biology, 2015).
Schneider, T.D. & Stephens, R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2015).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Hamming, R.W. Error detecting and error correcting codes. Bell Syst. Tech. J. 29, 147–160 (1950).
Huson, D.H. & Scornavacca, C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61, 1061–1067 (2012).
Rodriguez, R. et al. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 130, 15758–15759 (2008).
De Cian, A., Delemos, E., Mergny, J.-L., Teulade-Fichou, M.-P. & Monchaud, D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 129, 1856–1857 (2007).
Mailliet, P., Riou, J.-F., Mergny, J.-L., Laoui, A., Lavelle, F. & Petitgenet, O. Dérivés arylamines et leur application comme agent antitélomerase [Arylamine derivatives and their use as anti-telomerase agents]. Patent application WO 2001040218 (2001).
Acknowledgements
S.B. is a Wellcome Trust Senior Investigator (grant 099232/z/12/z). The Balasubramanian group is supported by European Research Council Advanced Grant 339778, and receives core (C14303/A17197) and program (C9681/A18618) funding from Cancer Research UK.
Author information
Authors and Affiliations
Contributions
All authors contributed to the concepts and design of the research. A.B.S. carried out the computational analyses. P.M. and C.M. carried out the retrotransposition experiments. All authors interpreted the data. A.B.S. and P.M. wrote the manuscript with contributions from all authors. S.B. supervised the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
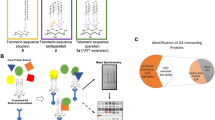
Supplementary Figure 1 34-nucleotide-long potential quadruplex sequences (PQSs) used for defining the L1-originated quadruplex sequence (LQS) family.
(a) Distribution of the Hamming distance of all 34-nt-long PQSs referenced against the most frequent PQS from L1 retrotransposons - LQSref. The peak with [0,5] borders, highlighted in red, defines the LQS family. The consensus sequence of the LQS family is illustrated via a sequence-logo plot. (b) Hierarchical clustering of all the 34-nt-long PQSs in the chromosome 1 with the Hamming distance applied as a similarity metrics. (c) Sub-trees depicting the region encompassing the LQS family (red box on panel b) together with some selected examples. (d) Genomic localization of repeat-associated PQSs and LQSs. It is noteworthy that almost all LQS sequences are found within the 3’-UTR of L1 elements. (e) Sequence composition of the LQS family of quadruplexes in different L1 subfamilies. Sequence-logo plots in e show the base frequencies at each LQS position in the retrotransposon remnants of a given subfamily. The analyzed subfamilies contain at least 5 conserved quadruplex sequences belonging to the LQS family.
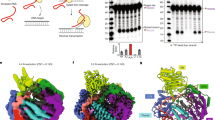
Supplementary Figure 2 The G-rich sequence found within the 3′ UTR of L1Hs folds into a G-quadruplex motif in vitro at both the DNA and the RNA level.
UV-melting profile followed at 295 nm of the L1Hs-DNA-G4 (a) and L1Hs-RNA-G4 (b) in the presence of 10 mM LiCl (orange line), NaCl (red line) or KCl (blue line). A cation-dependent hypochromic transition is characteristic of G4 formation. Circular dichroism (CD) spectra of the L1Hs-DNA-G4 (c) and L1Hs-RNA-G4 (d) in the presence of 100 mM LiCl (orange line), NaCl (red line) or KCl (blue line). All CD spectra are characterized by a minimum at 240 nm and a maximum at 263 nm characteristic of G4 formation. Expansion of the 1H NMR spectra of L1Hs-DNA-G4 (e) and L1Hs-RNA-G4 (f) pre-annealed in a 100 mM KCl containing buffer. Both NMR spectra exhibit characteristic imino proton signals shifted downfield (between 10 to 12 ppm) characteristic of Hoogsteen hydrogen bonding and G4 formation.
Supplementary Figure 3 Sanger sequencing traces for L1RPW, L1RPmG4 and L1RP∆G4 sequences.
The quadruplex motif found in the plasmid pL1RPWT (Addgene EF06R) was either mutated (pL1RPmG4) or deleted (pL1RPΔG4) by oe-PCR. Clones containing the desired plasmids were identified by Sanger sequencing.
Supplementary Figure 4 Biophysical characterization of the interaction of small-molecule G4 ligands with the DNA and RNA L1Hs quadruplexes.
Thermal denaturation profiles of L1HS-DNA-G4 (a) and L1HS-RNA-G4 (b) in the presence or absence of the small molecules PDS, PhenDC3, 12459, PDC12 and PDC20 followed by CD spectroscopy at 263 nm. The nucleic acids were annealed at 1.5 μM in a 5 mM and 1mM KCl containing buffer for the DNA or RNA quadruplex respectively. (c) Extracted melting temperatures (T1/2) from the CD melting experiments (data represent mean values ± s.d.; n = 3 technical replicates, i.e. independent melting experiments). (d) Concentration-dependent stabilization effect of the small molecules on Fl-L1HS-DNA-G4 (a dual fluorescently labeled L1HS DNA quadruplex) followed by FRET melting experiments (data represent mean values ± s.d.; n = 3 technical replicates). (e-f) Thermal denaturation profiles and extracted melting temperatures (T1/2) of Fl-L1Hs-DNA-G4 in the presence of PDC12 and an increasing concentration of a double-stranded DNA competitor (dsDNA). Fl-L1Hs-DNA-G4 was annealed at 100 nM in a 5 mM KCl containing buffer. All molecules, except PDC20, were found to stabilize both the DNA and RNA L1Hs quadruplexes. The newly reported compound PDC12 was found to be selective for the L1Hs-DNA quadruplex over double-stranded DNA (data in f represent mean values ± s.d.; n = 3 technical replicates).
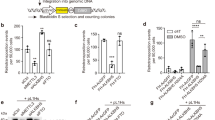
Supplementary Figure 5 Small-molecule G4 ligands stimulate retrotransposition of the human L1RP element in HeLa cells.
(a) Growth inhibition properties of the small molecules. GI50 values of growth inhibition were determined using the cell viability assay CellTiter-GloTM after 4 days of incubation (data represent mean values ± s.d.; n = 3 independent cell cultures). (b) Representative FACS profiles of HeLa cells transfected with either pL1RPWT or pL1RP∆G4 and treated with the different quadruplex ligands PDS, PhenDC3, 12459 and PDC12. PDC20 and HgS were used as negative and positive controls respectively. (c) Percentage of EGFP positive cells for the different treatments. The blue and red lines report the mean values of the percentages of EGFP positive cells transfected with pL1RPWT or pL1RP∆G4 mock treated with DMSO. (d) Percentage of EGFP positive cells fold changes compared to mock treatment. (e-g) The small molecule PDC12 stimulates the retrotransposition of the human L1RP element in HeLa cells in a concentration dependent manner. (e) Representative FACS profiles of HeLa cells transfected with either L1RPWT or L1RP∆G4 and treated with an increasing concentration of PDC12. (f) Percentage of EGFP positive cells for different doses of PDC12. The blue and red lines report the mean values of the percentages of EGFP positive cells transfected with L1RPWT or L1RP∆G4 in the absence of PDC12 respectively. (g) EGFP positive cells fold changes compared to mock treatment (DMSO) for different doses of PDC12. The individual frames in c, d, f and g represent the mean values ± s.d. for n = 3 independent experiments.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–3 and Supplementary Notes 1–2 (PDF 1981 kb)
Rights and permissions
About this article
Cite this article
Sahakyan, A., Murat, P., Mayer, C. et al. G-quadruplex structures within the 3′ UTR of LINE-1 elements stimulate retrotransposition. Nat Struct Mol Biol 24, 243–247 (2017). https://doi.org/10.1038/nsmb.3367
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3367
This article is cited by
-
Template and target-site recognition by human LINE-1 in retrotransposition
Nature (2024)
-
Telomeric retrotransposons show propensity to form G-quadruplexes in various eukaryotic species
Mobile DNA (2023)
-
LINE-1 regulates cortical development by acting as long non-coding RNAs
Nature Communications (2023)
-
Interaction between non-coding RNAs, mRNAs and G-quadruplexes
Cancer Cell International (2022)
-
Short single-stranded DNAs with putative non-canonical structures comprise a new class of plasma cell-free DNA
BMC Biology (2021)