Abstract
The current Zika virus (ZIKV) epidemic is characterized by severe pathogenicity in both children and adults. Sequence changes in ZIKV since its first isolation are apparent when pre-epidemic strains are compared with those causing the current epidemic. However, the residues that are responsible for ZIKV pathogenicity are largely unknown. Here we report the cryo-electron microscopy (cryo-EM) structure of the immature ZIKV at 9-Å resolution. The cryo-EM map was fitted with the crystal structures of the precursor membrane and envelope glycoproteins and was shown to be similar to the structures of other known immature flaviviruses. However, the immature ZIKV contains a partially ordered capsid protein shell that is less prominent in other immature flaviviruses. Furthermore, six amino acids near the interface between pr domains at the top of the spikes were found to be different between the pre-epidemic and epidemic ZIKV, possibly influencing the composition and structure of the resulting viruses.
Similar content being viewed by others
Main
The current ZIKV epidemic, which started in Brazil in 2015, has given rise to reports of microcephaly in fetuses1 and Guillain-Barre syndrome2 in adults. ZIKV is a member of the Flaviviridae family and, like some other flaviviruses, is transmitted by mosquitos. The flaviviruses include other significant human pathogens such as dengue virus (DENV), West Nile virus (WNV), yellow fever virus and Japanese encephalitis virus3. Flaviviruses are enveloped viruses containing a single-stranded, positive-sense RNA genome of ∼11 kb (ref. 3). In virions, the RNA genome complexes with multiple capsid protein molecules, surrounded by a membrane4. The extracellular, mature form of the virus has an outer surface that forms an icosahedral shell with 90 dimers of the envelope (E; ∼495 amino acids) and membrane (M; ∼75 amino acids) proteins, arranged in a herringbone pattern5. The E glycoprotein is involved in receptor binding, attachment and virus fusion during cell entry4. The M protein is mostly unexposed and has a significant transmembrane component. The cryo-EM structures of various mature flaviviruses5,6,7, including ZIKV8,9, are known and have a similar overall architecture (Fig. 1a,b). During infection of host cells, virions first assemble into an immature form of the virus in the endoplasmic recticulum3. The immature virus is composed of 60 trimeric spikes of the precursor membrane (prM) and E proteins; the pr domain (∼90 amino acids) of the prM protein protects the fusion loop on the E protein from nonproductive interactions within the cell10. In the low-pH environment of the trans-Golgi network, immature virions undergo proteolytic processing by furin, a cellular protease11, and the pr domain is cleaved from the prM protein during maturation. The virus then releases the pr domain to form the mature virion on exit from cells12.
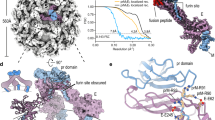
Surface view (a,c) and cross-section (b,d) of mature (top)8 and immature ZIKV (bottom), colored radially according to the keys. Thick black arrows indicate the densities between the inner RNA core and the viral membrane (double-ended arrow indicates the inside and outside layers of the membrane) in immature ZIKV. The numbered, thin black arrows in b and d give the icosahedral symmetry axes. The asymmetric unit is shown as a black triangle in a and c. Scale bar, 100 Å.
Although only the mature form of the virus is considered infectious, because of the varying efficiency of pr cleavage in flaviviruses3, the virus population secreted from host cells is a mixture of mature, partially mature and immature virions. Immature forms of flaviviruses such as DENV (ref. 13) and WNV (ref. 14) can become infectious through antibody-dependent entry into host cells. ZIKV is no exception to this observation of a mixed population of virus maturation states on release from infected cells. It is therefore probable that the immature form of ZIKV also plays a role in virus infection and spread.
Here we report a cryo-EM structure of the immature ZIKV (H/PF/2013 strain of Asian lineage) at a resolution of 9 Å. This structure has been used to fit the crystal structures of the prM and E proteins into the cryo-EM map and compared with immature virion structures from other flaviviruses.
Results
Cryo-EM structure of immature ZIKV
We infected mosquito C6/36 cells with ZIKV (strain H/PF/2013) for 16 h at 30 °C, and then added NH4Cl to produce immature virions. The purified immature ZIKV was plunge-frozen on grids and examined using an FEI Titan Krios electron microscope with a Gatan K2 Summit detector. Although the purified viruses were predominantly spiky, as expected for immature particles, they appeared to have some heterogeneity. A total of 9,315 particles were selected and used to generate a cryo-EM map at a resolution of 9.1 Å based on the 0.143 Fourier shell correlation criterion (Supplementary Fig. 1).
The cryo-EM reconstruction of the immature ZIKV has a spiky appearance with a diameter of approximately 600 Å (Fig. 1c,d). Densities for the individual prM–E heterodimers within the spikes are clearly visible in the immature ZIKV cryo-EM map. In addition, the individual transmembrane regions of the E and M proteins are easily recognizable. The crystal structure of the DENV-2 prM–E ectodomain heterodimer (PDB 3C6E)15 was fitted into the immature ZIKV cryo-EM map at the three independent positions with one trimeric spike (Supplementary Fig. 2a–c and Online Methods). The transmembrane helices of the M and E proteins from the mature ZIKV were also fitted individually into the transmembrane densities in the immature ZIKV cryo-EM map (Supplementary Fig. 2d). A comparison of these fitted components in ZIKV to the complete trimeric prM–E spike of immature DENV-1 (ref. 16) showed little difference in spatial arrangement.
Glycosylation in immature ZIKV
The fitting procedure showed that Asn69 of the pr domain in ZIKV is glycosylated, as it is in DENV (Supplementary Fig. 3a), but the glycosylation at Asn67 in the DENV E protein is not preserved in ZIKV (Supplementary Fig. 3b). Additional density was also found associated with each of the three Asn154 residues in immature ZIKV (Supplementary Fig. 3c), showing that Asn154 is glycosylated as expected from the protein sequence. Similarly, Asn154 on the E protein was glycosylated in the mature ZIKV and was found to cover the fusion loop in the adjacent E monomer, suggesting that this site maybe important for viral entry8.
Inner shell of capsid protein in immature ZIKV
We observed residual density between the inner membrane layer and the RNA core (Fig. 1d). The density is immediately below the base of each of the spikes and has a volume and shape that matches that of dimeric capsid protein structure17,18 (Figs. 1d and 2a–d). This potential capsid density is more evident in immature ZIKV than in immature DENV-1 (ref. 16). Mature flavivirus structures5,8 lack density in this region (Fig. 1b), suggesting a rearrangement of the capsid shell during maturation. The capsid density is in contact with the inner layer of the viral membrane and is close to the transmembrane domains of the E and M proteins, suggesting interactions that might be essential during virion assembly. The formation of an inner shell of capsid proteins also provides a benefit for recruitment of the viral genome. Moreover, the advantage of having a somewhat unstable inner core, as shown here for ZIKV, could be that it facilitates transfer of the genome into a host cell during infection.
(a) Cross-section of immature ZIKV projected down an icosahedral two-fold axis. The putative location of the inner capsid shell lies between the dashed concentric circles. Scale bar, 100 Å. (b,c) Magnified view of the region outlined by the red rectangle in a, showing the fitted NMR spectroscopy structure of the DENV-2 capsid protein (PDB 1R6R). Panel b is rendered at the same contour level as a (2.5), whereas c is rendered at a higher contour level (3.5), showing that the capsid protein density height is only about half of that of the outer glycoproteins. (d) Magnified view of the region outlined by the green rectangle in b showing the fitted DENV-2 capsid protein structure along two orthogonal views. Scale bars in b–d, 25 Å.
Interactions between prM–E heterodimers of the trimeric spikes
The three prM–E heterodimers that form a spike in immature flaviviruses are not related by threefold symmetry. Thus the interactions of any one of the three heterodimers with the two others in a spike is different. Our structure shows that the trimeric spike in immature ZIKV is held together at its external tip through interactions between the pr domains and the fusion loop of one of the E proteins (Fig. 3a,b). The residues involved in these interactions between prM–E heterodimers within the spikes can be deduced from the DENV-2 prM–E crystal structure fitted into the trimeric spike density of ZIKV, and the sequence alignment of prM–E for DENV-2 and ZIKV (H/PF/2013) (Fig. 3b, Supplementary Fig. 4 and Supplementary Table 1). Apart from these interactions at the top of the trimeric spikes, the base of the spike is stabilized by interactions between residues in domain III of the ZIKV E protein in one spike to the domain II of E protein from an adjacent spike (Fig. 3c, Supplementary Fig. 4 and Supplementary Table 1).
(a) Ribbon diagram of prM–E heterodimers in a trimeric spike of immature ZIKV. The prM–E heterodimers show E proteins in green, red and blue, whereas the pr domains are in brown, purple and black. Scale bar, 50 Å. (b) Diagram of the prM–E arrangement in a, using the same color-coding. The three prM–E molecules are marked 1, 2 and 3. The two different interaction regions within trimeric spikes are indicated by the gray dashed boxes. The molecule numbers and residues involved on either side of an interaction surface are shown in boxes colored according to the protein involved. (c) Diagrammatic representation of the trimeric spikes across adjacent asymmetric units. The interaction regions between E proteins from different spikes are indicated by gray dashed rectangles. A single trimeric spike is outlined in orange. The asymmetric unit is outlined by a black triangle.
Compared to the other viral proteins, the prM protein has the lowest percentage identity among flaviviruses (∼40%). Even when different strains of ZIKV are being compared, the pr domain has the highest percentage of changes in its amino acid sequence compared to other ZIKV proteins19. We noted six amino acid changes in the pr domain between pre-epidemic and epidemic ZIKV strains, and all of these were contained in the ∼40 amino acids at the start of the pr domain19 (Fig. 3). These amino acid changes clustered near the interface between pr domains and were involved in the interactions within trimeric spikes (Fig. 3b and Supplementary Fig. 4). A few of these amino acid substitutions involve dramatic changes in the polarity of the residues—for example, Lys21 and His35 in pre-epidemic ZIKV strains are substituted for by Glu21 and Tyr35 in the epidemic strains. Similarly, the change of Val26 to Pro26 between the pre-epidemic and epidemic ZIKV strains could potentially affect the local Cα backbone structure in the pr domain. Thus, these amino acid changes possess the ability to alter the nature of interactions within a trimer spike among the different strains of ZIKV.
Discussion
Comparison between the residues near the interface within trimeric spikes in DENV to those in the epidemic ZIKV strains showed that the interface in DENV is more positively charged than that in ZIKV (Supplementary Fig. 4). The lack of such closely positioned positively charged residues in ZIKV could imply comparatively more stable interactions within spikes in immature ZIKV. This would have an effect on the dynamics of conformational change between the immature and mature virion states, wherein the trimeric spikes have to break contacts to form the dimeric interactions seen in mature virions. Thus, ZIKV might tend to have a higher proportion of partially immature virions in their populations than of completely mature virions. This partial maturation of virions is seen in all flaviviruses, but the degree of heterogeneity of virion particles varies between flaviviruses3. A structurally heterogeneous virion population is advantageous to a viral pathogen, as it limits the uniform availability of neutralizing epitopes on the virion, making it more challenging for the host immune system to inhibit the virus. This phenomenon is observed not only with other flaviviruses, such as WNV20,21, but also for other debilitating viruses such as HIV-1 (refs. 22,23). In addition, non-neutralizing antibody responses can be counteractively used by ZIKV for antibody-dependent entry of host cells24,25. Thus, local differences between ZIKV strains could modulate the sensitivity of ZIKV to antibodies and impact the potency of viral infection.
Methods
Virus preparation and purification.
Approximately 1 × 109 C6/36 cells (ATCC CRL-1660) were infected with Zika virus (strain H/PF/2013) at a multiplicity of infection of 4 for 16 h at 30 °C. The inoculum was removed, and cells were rinsed three times with phosphate-buffered saline (pH 8.0) and incubated for 2 h at 30 °C in minimal essential media containing 2% FBS, 25 mM HEPES, pH 7.3, and 30 mM NH4Cl. The process was repeated two more times for a total of three incubations at 30 °C in cell culture media containing 30 mM NH4Cl. Media was collected 72 h after infection, and immature virus particles were purified according to previously described methods12. Briefly, virus particles were precipitated from the media by overnight mixing with 8% polyethylene glycol 8000 at 4 °C. This mixture was then pelleted at 8,900g for 50 min at 4 °C. Resuspended particles were pelleted through a 24% sucrose cushion, resuspended in 0.5 mL of NTE buffer (20 mM Tris, pH 8.0, 120 mM NaCl, 1 mM EDTA) and purified with a discontinuous gradient in 5% intervals from 35% to 10% potassium tartrate, 20 mM Tris, pH 8.0, and 1 mM EDTA. Immature virus was extracted from the gradient and concentrated, and the buffer was exchanged for NTE buffer.
Cryo-electron microscopy data collection.
The purified immature ZIKV sample was plunge-frozen on ultrathin lacey carbon EM grids (Electron Microscopy Sciences). The grids were examined using an FEI Titan Krios electron microscope with a Gatan K2 Summit detector. Cryo-EM images were collected at a magnification of 22,500× in the 'super-resolution' data collection mode. The pixel size of the collected images was 0.65 Å. The total exposure time for producing one image composed of 38 frames was 7.6 s and required a dose rate of 4.7 electrons Å−2 s−1. A total of 3,341 images were collected and 14,351 particles were boxed manually using the e2boxer program in EMAN2 (ref. 26).
Three-dimensional reconstruction and data analysis.
Nonreference two-dimensional classification was performed using the Relion software package27, resulting in the selection of 9,315 particles. This data set was split into two equal subsets as required by the 'gold standard' for determining the quality of the cryo-EM reconstruction. The JSPR program28 was used for initial model generation and refinement of the orientations of the selected particles. The selected particles were used to generate a cryo-EM map at an average resolution of 9.34 Å based on the 0.143 Fourier shell correlation criterion. After the application of soft spherical masks, the resolution improved to 9.14 Å.
Fitting of crystal structures into the cryo-EM map.
The crystal structure of the immature DENV-2 prM–E heterodimer (PDB 3C6E) was used to fit a trimeric spike in the immature ZIKV map. Sequential fitting of the three prM–E molecules into the trimeric spike density was carried out using UCSF Chimera software29. The Cα backbone of the transmembrane helices of M and E proteins from the mature ZIKV structure (PDB 5IRE) were also fitted into the transmembrane regions of the three prM–E heterodimers. The solution structure of DENV-2 capsid protein (PDB 1R6R) was placed into the density between the inner layer of the viral membrane and the RNA core of immature ZIKV, according to a previously suggested orientation of the capsid protein in the virion17. The ectodomain structure (PDB 5IRE)8 of mature ZIKV E protein was split into two parts, with one molecule containing domains I and III and the second molecule containing domain II. These different parts were superposed independently on the immature dengue prM–E molecule to determine the interaction regions between E proteins from adjacent spikes. Interface regions between different protein partners were identified as residues with less than 6 Å distance between their corresponding Cα backbones.
Multiple sequence alignment.
Sequence alignments of the pr domains and E glycoproteins among flaviviruses were carried out using the MAFFT program30 and rendered using ESPript software31. The sequences for comparison were obtained from the ViPR resource database32. The representative flavivirus strains used in the comparison were Zika-Asian (H/PF/2013 strain), Zika-African (1968-Nigeria strain), West Nile virus (NY99 strain), Japanese encephalitis (SA14 strain), yellow fever (Asibi strain) and the four dengue virus serotypes (Western Pacific, S16803, CH53489 and IND1979 strains). In this study, the residue numbers for pr and E used were chosen on the basis of the ZIKV sequence and structure, though the numbers assigned to the residues during multiple sequence alignment may be slightly shifted in Supplementary Figure 4.
Data availability.
The atomic coordinates of the fitted dengue virus-2 prM–E molecules and mature ZIKV E and M transmembrane components along with the cryo-EM density map of the immature ZIKV are available at the Protein Data Bank and Electron Microscopy Data Bank with accession codes 5U4W and EMD-8508, respectively.
References
Schuler-Faccini, L. et al. Possible association between Zika virus infection and microcephaly - Brazil, 2015. MMWR Morb. Mortal. Wkly. Rep. 65, 59–62 (2016).
Chan, J.F., Choi, G.K., Yip, C.C., Cheng, V.C. & Yuen, K.Y. Zika fever and congenital Zika syndrome: An unexpected emerging arboviral disease. J. Infect. 72, 507–524 (2016).
Lindenbach, B.D., Murray, C.L., Thiel, H.-J. & Rice, C.M. Fields Virology (eds. Knipe, D.M. et al.) (Lippincott Williams & Wilkins, 2013).
Mukhopadhyay, S., Kuhn, R.J. & Rossmann, M.G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3, 13–22 (2005).
Kuhn, R.J. et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108, 717–725 (2002).
Mukhopadhyay, S., Kim, B.S., Chipman, P.R., Rossmann, M.G. & Kuhn, R.J. Structure of West Nile virus. Science 302, 248 (2003).
Zhang, X. et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 20, 105–110 (2013).
Sirohi, D. et al. The 3.8 Å resolution cryo-EM structure of Zika virus. Science 352, 467–470 (2016).
Kostyuchenko, V.A. et al. Structure of the thermally stable Zika virus. Nature 533, 425–428 (2016).
Zhang, Y. et al. Structures of immature flavivirus particles. EMBO J. 22, 2604–2613 (2003).
Stadler, K., Allison, S.L., Schalich, J. & Heinz, F.X. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71, 8475–8481 (1997).
Yu, I.-M. et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319, 1834–1837 (2008).
Rodenhuis-Zybert, I.A. et al. Immature dengue virus: a veiled pathogen? PLoS Pathog. 6, e1000718 (2010).
Colpitts, T.M. et al. prM-antibody renders immature West Nile virus infectious in vivo. J. Gen. Virol. 92, 2281–2285 (2011).
Li, L. et al. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319, 1830–1834 (2008).
Kostyuchenko, V.A., Zhang, Q., Tan, J.L., Ng, T.S. & Lok, S.M. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J. Virol. 87, 7700–7707 (2013).
Ma, L., Jones, C.T., Groesch, T.D., Kuhn, R.J. & Post, C.B. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. USA 101, 3414–3419 (2004).
Dokland, T. et al. West Nile virus core protein; tetramer structure and ribbon formation. Structure 12, 1157–1163 (2004).
Wang, L. et al. From mosquitos to humans: genetic evolution of Zika virus. Cell Host Microbe 19, 561–565 (2016).
Pierson, T.C. et al. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe 1, 135–145 (2007).
Nelson, S. et al. Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog. 4, e1000060 (2008).
Davenport, T.M. et al. Isolate-specific differences in the conformational dynamics and antigenicity of HIV-1 gp120. J. Virol. 87, 10855–10873 (2013).
Kwong, P.D. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002).
Dejnirattisai, W. et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat. Immunol. 17, 1102–1108 (2016).
Tirado, S.M. & Yoon, K.J. Antibody-dependent enhancement of virus infection and disease. Viral Immunol. 16, 69–86 (2003).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Scheres, S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Guo, F. & Jiang, W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol. 1117, 401–443 (2014).
Pettersen, E.F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Katoh, K. & Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Pickett, B.E. et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 40, D593–D598 (2012).
Acknowledgements
We thank M. Sevvana for discussions about the manuscript. We thank S. Kelly for helping us prepare the manuscript. We also thank the Purdue Cryo-EM Facility for equipment access and support. This work was supported by the National Institutes of Health (RO1 AI076331 to M.G.R. and R.J.K., and a subaward for RO1 AI073755 (principal investigator: M.S. Diamond, Washington University) to both M.G.R. and R.J.K.).
Author information
Authors and Affiliations
Contributions
A.S.M., D.S. and G.B. were involved in preparation of cell culture, and optimization and purification of virus sample; V.M.P. and T.K. conducted the cryo-EM preparation, data collection and data processing; V.M.P. performed the data analyses; W.J. made his JSPR program available for reconstruction and refinement of the cryo-EM map; and V.M.P., R.J.K. and M.G.R. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Resolution estimation using Fourier shell correlation (FSC).
A plot of FSC against resolution in Å-1, calculated using two independent half-sets of the cryo-EM data. The resolution of the map is 9.1 Å using the 0.143 FSC cut-off (light blue line).
Supplementary Figure 2 Fit of the DENV-2 prM–E crystal structure into the immature ZIKV cryo-EM map.
a, Surface view of the complete immature ZIKV. One of the 60 trimeric spikes is identified in color, with its E proteins in green and the pr domains in orange. The asymmetric unit is shown as a black triangle. The scale bar is 100 Å long. b, Top view showing the fit of three prM-E heterodimers into the cryo-EM spike density. The three independent prM-E heterodimers are colored in light green, blue and orange for the E proteins and dark green, dark blue and red for the pr domains, respectively. c, Side-view of the fitted prM-E heterodimers in a spike. The dashed black line shows the position of the viral membrane surface. d, Cryo-EM density showing the individual fitting of the transmembrane regions of E (blue) and M (magenta) proteins. Scale bar is 50 Å long in b, c and d.
Supplementary Figure 3 Analysis of crystal structures fitted into immature ZIKV cryo-EM map.
a, Cryo-EM density showing the conservation of Asn-69 glycosylation site in the pr domains of immature ZIKV. The DENV-2 prM-E crystal structure (PDB ID: 3C6E) including the Asn-69 glycan has been fitted into all three positions within a trimeric spike. The glycans attached to Asn-69 are shown in blue. Scale bar is 25 Å in length. b, Glycan (colored in blue) attached to Asn-67 in the DENV-2 E protein structure and fitted into the map of the immature ZIKV showing the absence of density associated with the glycan at this site in ZIKV. c, Additional density adjacent to Asn-154 (red) in immature ZIKV indicating the presence of a glycan. Scale bar is 10 Å long in b and c. Black arrows in a and c point to the densities of the glycans. The E proteins and pr domains are shown in green and orange respectively in all panels.
Supplementary Figure 4 Alignment of sequences that form the interfaces between immature ZIKV spikes.
Top panel, Alignment of pr domain sequences from different flaviviruses. The residues that differ between the pre-epidemic (African) and epidemic (Asian) ZIKV strains are indicated in blue boxes. Bottom panel, Alignment of parts of the E protein sequence from different flaviviruses. In both panels, red columns indicate strictly conserved residues, yellow columns show partially conserved residues and white columns show variable residues. Residues involved in interactions within a trimeric spike and between adjacent spikes are indicated by brown and green colored bars above the corresponding residues, respectively. The furin cleavage site at the end of the pr domain is shown by a grey bar. Abbreviations DEN, JEV and YFV refer to dengue virus, Japanese encephalitis virus and yellow fever virus respectively.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Table 1 (PDF 1060 kb)
Rights and permissions
About this article
Cite this article
Prasad, V., Miller, A., Klose, T. et al. Structure of the immature Zika virus at 9 Å resolution. Nat Struct Mol Biol 24, 184–186 (2017). https://doi.org/10.1038/nsmb.3352
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3352
This article is cited by
-
Zika virus prM protein contains cholesterol binding motifs required for virus entry and assembly
Nature Communications (2023)
-
Zika virus M protein latches and locks the E protein from transitioning to an immature state after prM cleavage
npj Viruses (2023)
-
Adaptation to host cell environment during experimental evolution of Zika virus
Communications Biology (2022)
-
Construction of a recombinant avipoxvirus expressing the env gene of Zika virus as a novel putative preventive vaccine
Virology Journal (2021)
-
Leu-to-Phe substitution at prM146 decreases the growth ability of Zika virus and partially reduces its pathogenicity in mice
Scientific Reports (2021)