Abstract
Nucleosomes are essential for proper chromatin organization and the maintenance of genome integrity. Histones are post-translationally modified and often evicted at sites of DNA breaks, facilitating the recruitment of repair factors. Whether such chromatin changes are localized or genome-wide is debated. Here we show that cellular levels of histones drop 20–40% in response to DNA damage. This histone loss occurs from chromatin, is proteasome-mediated and requires both the DNA damage checkpoint and the INO80 nucleosome remodeler. We confirmed reductions in histone levels by stable isotope labeling of amino acids in cell culture (SILAC)-based mass spectrometry, genome-wide nucleosome mapping and fluorescence microscopy. Chromatin decompaction and increased fiber flexibility accompanied histone degradation, both in response to DNA damage and after artificial reduction of histone levels. As a result, recombination rates and DNA-repair focus turnover were enhanced. Thus, we propose that a generalized reduction in nucleosome occupancy is an integral part of the DNA damage response in yeast that provides mechanisms for enhanced chromatin mobility and homology search.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Boettiger, A.N. et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 (2016).
Aze, A., Sannino, V., Soffientini, P., Bachi, A. & Costanzo, V. Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nat. Cell Biol. 18, 684–691 (2016).
Gerhold, C.B., Hauer, M.H. & Gasser, S.M. INO80-C and SWR-C: guardians of the genome. J. Mol. Biol. 427, 637–651 (2015).
Seeber, A., Hauer, M. & Gasser, S.M. Nucleosome remodelers in double-strand break repair. Curr. Opin. Genet. Dev. 23, 174–184 (2013).
Soria, G., Polo, S.E. & Almouzni, G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol. Cell 46, 722–734 (2012).
Chiolo, I. et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144, 732–744 (2011).
Lemaître, C. et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 28, 2450–2463 (2014).
Torres-Rosell, J. et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 9, 923–931 (2007).
Dion, V., Kalck, V., Horigome, C., Towbin, B.D. & Gasser, S.M. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 14, 502–509 (2012).
Miné-Hattab, J. & Rothstein, R. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 14, 510–517 (2012).
Roukos, V. et al. Spatial dynamics of chromosome translocations in living cells. Science 341, 660–664 (2013).
Seeber, A., Dion, V. & Gasser, S.M. Checkpoint kinases and the INO80 nucleosome remodeling complex enhance global chromatin mobility in response to DNA damage. Genes Dev. 27, 1999–2008 (2013).
Hu, B. et al. Biological chromodynamics: a general method for measuring protein occupancy across the genome by calibrating ChIP-seq. Nucleic Acids Res. 43, e132 (2015).
Povirk, L.F., Wübter, W., Köhnlein, W. & Hutchinson, F. DNA double-strand breaks and alkali-labile bonds produced by bleomycin. Nucleic Acids Res. 4, 3573–3580 (1977).
Gunjan, A. & Verreault, A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115, 537–549 (2003).
Liang, D., Burkhart, S.L., Singh, R.K., Kabbaj, M.H. & Gunjan, A. Histone dosage regulates DNA damage sensitivity in a checkpoint-independent manner by the homologous recombination pathway. Nucleic Acids Res. 40, 9604–9620 (2012).
Heinemeyer, W., Kleinschmidt, J.A., Saidowsky, J., Escher, C. & Wolf, D.H. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 10, 555–562 (1991).
Dion, V. & Gasser, S.M. Chromatin movement in the maintenance of genome stability. Cell 152, 1355–1364 (2013).
Krawczyk, P.M. et al. Chromatin mobility is increased at sites of DNA double-strand breaks. J. Cell Sci. 125, 2127–2133 (2012).
Strecker, J. et al. DNA damage signalling targets the kinetochore to promote chromatin mobility. Nat. Cell Biol. 18, 281–290 (2016).
Celona, B. et al. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol. 9, e1001086 (2011).
Sanchez, Y. et al. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271, 357–360 (1996).
Morrison, A.J. et al. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell 130, 499–511 (2007).
Poli, J. et al. Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev. 30, 337–354 (2016).
Lafon, A. et al. INO80 chromatin remodeler facilitates release of RNA polymerase II from chromatin for ubiquitin-mediated proteasomal degradation. Mol. Cell 60, 784–796 (2015).
Chen, X. et al. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489, 576–580 (2012).
van Attikum, H., Fritsch, O. & Gasser, S.M. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26, 4113–4125 (2007).
Lisby, M., Barlow, J.H., Burgess, R.C. & Rothstein, R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 (2004).
Verdaasdonk, J.S. et al. Centromere tethering confines chromosome domains. Mol. Cell 52, 819–831 (2013).
Haase, S.B. & Lew, D.J. Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol. 283, 322–332 (1997).
Liu, C., Apodaca, J., Davis, L.E. & Rao, H. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques 42, 158–162 (2007).
Lee, D.H. & Goldberg, A.L. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 271, 27280–27284 (1996).
Wiechens, N. et al. The chromatin remodelling enzymes SNF2H and SNF2L position nucleosomes adjacent to CTCF and other transcription factors. PLoS Genet. 12, e1005940 (2016).
Gruhler, A. et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 (2005).
Pasero, P., Duncker, B.P., Schwob, E. & Gasser, S.M. A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev. 13, 2159–2176 (1999).
Sage, D., Neumann, F.R., Hediger, F., Gasser, S.M. & Unser, M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans. Image Process. 14, 1372–1383 (2005).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Dietz, C. & Berthold, M.R. KNIME for open-source bioimage analysis: a tutorial. Adv. Anat. Embryol. Cell Biol. 219, 179–197 (2016).
Sommer, C. & Gerlich, D.W. Machine learning in cell biology—teaching computers to recognize phenotypes. J. Cell Sci. 126, 5529–5539 (2013).
Acknowledgements
M.H.H. thanks S. Koren-Hauer for critical reading and assistance in preparing the manuscript, and V. Dion and H. Ferreira for fruitful discussions and advice. We thank V. Dion (Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland; strain GA-5816), J. Haber (Department of Biology and Rosenstiel Medical Center, Brandeis University, Waltham, Massachusetts, USA; strain JKM-179), B. Luke (Institute of Molecular Biology, Mainz, Germany; strain GA-3364), K. Bystricky (University of Toulouse, Toulouse, France; precursor strain for GA-9777, strain GA-9227), J.-M. Galan (Institut Jacques Monod, Paris, France; strains GA-1364, GA-1365 and GA-1366) and F. Winston (Department of Genetics, Harvard Medical School, Boston, Massachusetts, USA; plasmids 3494 and 3495) for reagents and material. We are grateful for the technical assistance provided by L. Gelman (microscopy), S. Bourke (microscopy) and H. Kohler (FACS). M.H.H. thanks the Bioinformatics facility for help in getting started with R. We thank all members of the FMI Protein Analysis and Microscopy facilities for valuable advice and support. We thank all members of the Gasser laboratory for valuable discussions and technical support. M.H.H. was supported by a PhD fellowship of the Boehringer Ingelheim Fonds. S.M.G. thanks the HFSP, SNSF and the Novartis Research Foundation for support.
Author information
Authors and Affiliations
Contributions
M.H.H. and S.M.G. wrote the manuscript. M.H.H. designed experiments and analyzed the data. M.H.H. performed most of the experiments. A.S. contributed to experimental design, data analysis and writing of the manuscript, and performed high-speed live-cell tracking after Zeocin treatment. M.H.H. planned and M.H.H. and A.S. performed the ectopic integration assays and the Rad52-YFP recovery assay. M.K. assisted in ectopic recombination assays. V.S. and T.O.-H. performed and analyzed genome-wide nucleosome mapping. A.A. and D.H. performed biophysical analysis of high-speed tracking data. R.T. performed and maintained the coding for 3D SIM-data analysis. R.S. performed all mass spectrometry measurements and the analysis of label-free experiments. J.E. performed and maintained the coding of tools for 3D interspot distance measurements. All the authors discussed the data and participated in the preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
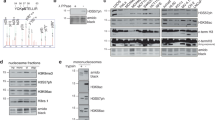
Supplementary Figure 1 SILAC mass spectrometry of pre-enriched chromatin depicts core histone loss.
(a) Experimental workflow for SILAC mass spectrometry after Zeocin treatment. (b) Labeling and mixing of samples from 4 individual experiments. Asterisks indicates label swap (c) Colloidal Commassie stained SDS-PAGE of SILAC experiment replicas showing total protein, supernatant (SUP), and chromatin (CHR) fractions from a. His. mix is an equimolar mixture of recombinant Histone H2A, H2B, H3 and H4. (d) Control Immunoblot analysis using anti-γH2A anti-Rad53 antibodies to show that checkpoint is activated after Zeocin treatment in the SILAC samples from bc. (e) FACS analysis showing that all samples from b-d have similar cell cycle profiles. Actin was used as loading control. Asterisks indicate the phosphorylation-dependent mobility shift of Rad53. (f) SILAC mass spectrometry on chromatin fractions from three independent cell pools. Boxplots show heavy/light histone peptide distribution indicating the degradation of core histones and, to a lesser extent, Htz1 (H2A.Z). (g) Distribution of measured protein ratios in the non-label swap experiment or (h) label swap experiment. Core histones are labelled red and reside within the μ-σ range. Htz1 is labelled yellow and resides closer to the mean ratio of all proteins. Boxplots in f represent median values, interquartile ranges and whiskers.
Supplementary Figure 2 Gamma irradiation triggers degradation of core histones, Zeocin reduces nucleosome occupancy, and H2B-CFP tagging does not interfere with cell viability.
(a) Immunoblot analysis from one experiment using H3 and H4 specific antibodies on whole cell extracts of asynchronous WT cells exposed to 30 Gy gamma irradiation (γ-IR). Rad53 and γH2A were probed to confirm checkpoint activation. MCM2 was used to control for loading. Arrows indicate samples sent for label-free quantitative mass spectrometric analysis. (b) Label-free quantitative mass spectrometry results of samples depicted in a. Bar graphs show mean peptide ratios ± s.e.m for the indicated histone proteins upon γIR exposure relative to the control condition. (c) Combined label-free mass spectrometry results of sample γ-IR a), γIR b) and an additional experiment. Bar graphs represent the mean peptide ratios (γIR/Ctr.) ± s.e.m. for core histones over all samples. (d) FACS analysis showing that all samples have similar cell cycle profiles. (e) Genome-wide nucleosome mapping graph shows the distribution of nucleosome reads over 750 highly expressed genes aligned to their TSS from four independent experiments (±s.d. is shaded). (f) Drop assay control showing that the H2B-CFP fusion complements the absence of H2B in response to genotoxic agents. (g) Live single-cell microscopy of Nup49-GFP. Graph shows the the mean fluorescent signals of of all individual cells (cell numbers indicated in graph) per treatment over time relative to the control (Ctr.) condition. Dotted lines indicate the duration of Zeocin treatment. Graphs show mean ± s.e.m..
Supplementary Figure 3 Damage-induced histone loss occurs in G1 phase.
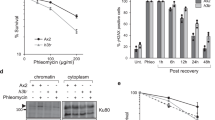
(a-b) Representative immunoblot analysis of whole cell extracts from G1-arrested cells treated with Zeocin a or after exposure to γIR b. Histone H3 and H4 levels were probed using histone specific antibodies. Rad53 and γH2A were probed to confirm checkpoint activation. MCM2 was used to control for loading and Ctr. represents bands on the ponceau stained membrane. Bar graphs in a show the mean ± s.e.m. over three independent replicates relative to the control condition. FACS results of Zeocin treated samples are shown above immunoblots in a. Arrows in b indicate samples sent for label-free quantitative mass spectrometric analysis. (c) Immunoblot quantifications of irradiated samples from one experiment marked with arrows. (d) Label-free quantitative mass spectrometry results of samples depicted with arrows. Bar graphs show mean peptide ratios ± s.e.m. for the indicated histone proteins upon γIR exposure relative to the control condition. (e) FACS analysis showing cell cycle profiles of all samples from b. (f) Experimental workflow for SILAC mass spectrometry of G1 arrested cells after Zeocin treatment. (g) Commassie stained SDS-PAGE of samples showing total protein, supernatant (SUP), and chromatin (CHR) fractions. (h) FACS analysis showing similar G1 arrest efficiency for all samples. (i) SILAC mass spectrometry on chromatin fractions from two independent cells pools. Boxplots show heavy/light histone peptide distribution indicating the degradation of core histones and, to a lesser extent, Htz1. (j) Distribution of measured proteins ratios. Core histones are labelled red and reside within the μ-σ range. Htz1 is labelled yellow and residues closer to the mean ratio of all proteins. Boxplots in i represent median values, interquartile ranges and whiskers. Asterisk indicates phosphorylation-dependent Rad53 mobility shift.
Supplementary Figure 4 Damage-induced histone loss is independent of histone transcription.
(a) Top panel shows the experimental procedure and strain used for constitutive histone H3 and H4 transcription in cells grown in YPGal:Raff medium (strain GA-8386). A URA3 plasmid borne construct in which the GAL1-10 promoter drives the only pair of histone H3/H4 genes is used. Mid panel shows representative immunoblot analysis using anti-H3 and anti-H4 antibodies on whole cell extracts from the strain depicted in a after Zeocin treatment and growth in YPGal:Raff medium. Rad53 and γH2A were probed to confirm checkpoint activation. MCM2 was used to control for loading and Ctr. represents bands on the original gel (UV-TGX stained). Bar graphs in bottom panel show the mean ± s.e.m. over three independent replicates relative to the control condition. Asterisk indicates phosphorylation-dependent Rad53 mobility shift. (b) Zeocin treatment causes a genome-wide decrease in nucleosome occupancies. Data represents nucleosome occupancies over the total pool of 5014 protein coding genes, 750 high expression genes and 750 low expression genes aligned to their transcriptional start site (TSS) from one experiment using the strain depicted in a.
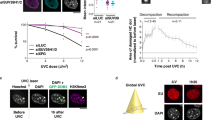
Supplementary Figure 5 High-speed, live-cell imaging reveals increased chromatin movement and a loss of constraining forces after DNA damage.
(a) High-speed (Δt=80 ms) imaging of the undamaged MET10 locus (as in Fig. 3a-b) showing that chromatin mobility increases with Zeocin concentration. Average MSD graphs indicate dose-dependent increases in global chromatin mobility in response to DNA damage (nCtr.=39, nZeo200=31, nZeo500=29 different cells from three independent experiments). (b) Graphs show the means and whiskers (±s.d.) of biophysical parameters derived from imaging data and predict chromatin decompaction after Zeocin treatment. P-values, ***P<0.001, NS=not significant, result from Kolmogorov-Smirnov-tests. All MSD graphs represent the mean ± s.e.m. of cells pooled from three independent experiments. Additionally, consult Supplementary Dataset 2 for mobility parameters and the number of cells analyzed.
Supplementary Figure 6 GAL::H3/H4 strain as a tool for in vivo artificially controlled histone-level reductions.
(a) Schematic representation of wild-type, control and shutdown strains grown in the indicated media. Gal. = galactose, gluc. = glucose. Immunoblot analysis of whole cell extracts of the indicated conditions and strains were performed using an antibody directed against Histone H4. Rad53 and γH2A were probed to confirm checkpoint activation. MCM2 was used to control for loading. Bar graphs from quantified immunoblot derived from one experiment shows overexpression or reduction of H3/H4 in the shutdown strain grown in gal. or gluc. medium respectively. Growth of the shutdown strain in Gal:Raff 1:5 confers H3/H4 levels similar to WT. (b) Experimental workflow of the arrest-release experiment used to reduce histone levels in S phase (as in Fig. 5). Bar graphs from quantified immunoblot data derived from one experiment shows reductions of H3 and H4 upon release into raffinose medium. (c) A defined number of exponentially growing cells (fivefold dilutions) was spotted on different YP or YPD plates containing the indicated dose of hydroxyurea (HU). Cells exposed to 20 Gy γIR were spotted onto YPD plates. Drop assays show functionality of shutdown and control strains. Control = control from a, control 1 and 2 = similar to control 1 but expressing HHT2-HHF2 from a URA3 plasmid (a).
Supplementary Figure 7 Biophysical parameters of nhp6∆ tracking data and results from Rad52-YFP recovery assay.
(a) Control Immunoblot from one experiment (loading 1x and 2x the volume) showing that nhp6Δ strains do not have constitutive checkpoint activation. Rad53 was probed to test for checkpoint activation and MCM2 was used as loading control. (b) Schematics of the strains used for imaging the PES4 and MET10 loci (Fig. 6c-e) with representative images. Scale bar is 2 μm. (c-d) Graphs show the means and whiskers (±s.d.) of biophysical parameters derived from imaging data of PES4 c and MET10 d (Fig. 6c,d). P-values, *P<0.05, ***P<0.001, NS=not significant, result from Kolmogorov-Smirnov-Tests. (e) Rad52-YFP foci recovery assay. Graph shows the overall percentage of Rad52-YFP foci containing cells for each of the 12 time-points from one experiment plotted against the time and shown together with a logarithmic fit.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1–4 and Supplementary Note 1 (PDF 2179 kb)
Supplementary Dataset 1: Uncropped immunoblot images
Uncropped blot images used in Figs. 1b, 2a, 2b, 2c and 7a. (PDF 6366 kb)
Supplementary Dataset 2: Summary of mobility parameters
Table showing the strains, conditions and mobility parameters. (XLSX 27 kb)
Supplementary Dataset 3: KNIME workflow
File contains the KNIME workflow used for imaging data analysis. (ZIP 107 kb)
Supplementary Dataset 4: MS search results: peptides, cycling cells, label swap
File contains MaxQuant search results used for quantifications. (XLSX 1374 kb)
Supplementary Dataset 5: MS search results: protein groups, cycling cells, label swap
File contains MaxQuant search results used for quantifications. (XLSX 360 kb)
Supplementary Dataset 6: MS search results: peptides, cycling cells, non–label swap
File contains MaxQuant search results used for quantifications. (XLSX 1748 kb)
Supplementary Dataset 7: MS search results: protein groups, cycling cells, non–label swap
File contains MaxQuant search results used for quantifications. (XLSX 494 kb)
Supplementary Dataset 8: MS search results: peptides, G1 arrest cells
File contains MaxQuant search results used for quantifications. (XLSX 873 kb)
Supplementary Dataset 9: MS search results: protein groups, G1 arrest cells
File contains MaxQuant search results used for quantifications. (XLSX 243 kb)
H2B-CFP intensity decreases in response to DNA damage.
Visualization of data shown in Fig. 1d. Exemplary time course of 9 individual cells following H2B-CFP intensities after 60 min (20–80-min time point) treatment with 300 μg/ml Zeocin for a total time of 120 min. Shown is a merge of Brightfield (average intensity projections) and CFP (maximum intensity projection) channels. Time-lapse series (120 min total) of 100 optical slices per stack (200-nm intervals) were acquired for 12 time points at 10-min intervals, with each slice being exposed for 10 ms per laser line. Video was generated with Fiji (ImagJ) and is shown at 2 frames per second. Original Δt is shown in the top right corner. (MP4 542 kb)
CFP-LacI and TetR-mRFP time course used for live-cell 3D interdistance measurements.
Visualization of data shown in Fig. 4b. Exemplary time course of CFP-LacI and TetR-mRFP used for 3D interdistance measurements in living cells. The fluorescent channels were acquired simultaneously on two different CCD cameras, taking 8 optical slices (200-nm thickness) per stack every 300 ms for 2 min, with 10-ms exposure times per slice. Video was generated using the Imaris 8.2.0 software and is shown at 25 frames per second (7.5× faster than the original acquisition speed). (MP4 1330 kb)
Rights and permissions
About this article
Cite this article
Hauer, M., Seeber, A., Singh, V. et al. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat Struct Mol Biol 24, 99–107 (2017). https://doi.org/10.1038/nsmb.3347
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3347
This article is cited by
-
HJURP is recruited to double-strand break sites and facilitates DNA repair by promoting chromatin reorganization
Oncogene (2024)
-
Aedes aegypti Argonaute 2 controls arbovirus infection and host mortality
Nature Communications (2023)
-
Circulating DNA reveals a specific and higher fragmentation of the Y chromosome
Human Genetics (2023)
-
Cancer cell histone density links global histone acetylation, mitochondrial proteome and histone acetylase inhibitor sensitivity
Communications Biology (2022)
-
Transcription coordinates histone amounts and genome content
Nature Communications (2021)