Abstract
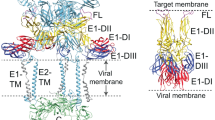
The lipid-enveloped influenza virus enters host cells during infection by binding cell-surface receptors and, after receptor-mediated endocytosis, fusing with the membrane of the endosome and delivering the viral genome and transcription machinery into the host cell. These events are mediated by the hemagglutinin (HA) surface glycoprotein. At the low pH of the endosome, an irreversible conformational change in the HA, including the exposure of the hydrophobic fusion peptide, activates membrane fusion. Here we used electron cryomicroscopy and cryotomography to image the fusion of influenza virus with target membranes at low pH. We visualized structural intermediates of HA and their interactions with membranes during the course of membrane fusion as well as ultrastructural changes in the virus that accompany membrane fusion. Our observations are relevant to a wide range of protein-mediated membrane-fusion processes and demonstrate how dynamic membrane events may be studied by cryomicroscopy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Skehel, J.J. & Wiley, D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569 (2000).
Wilson, I.A., Skehel, J.J. & Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289, 366–373 (1981).
Skehel, J.J. et al. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. USA 79, 968–972 (1982).
Ruigrok, R.W. et al. Electron microscopy of the low pH structure of influenza virus haemagglutinin. EMBO J. 5, 41–49 (1986).
Bullough, P.A., Hughson, F.M., Skehel, J.J. & Wiley, D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 (1994).
Ruigrok, R.W. et al. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J. Gen. Virol. 69, 2785–2795 (1988).
Chen, J. et al. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc. Natl. Acad. Sci. USA 92, 12205–12209 (1995).
Chen, J., Skehel, J.J. & Wiley, D.C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 96, 8967–8972 (1999).
Tsurudome, M. et al. Lipid interactions of the hemagglutinin HA2 NH2-terminal segment during influenza virus-induced membrane fusion. J. Biol. Chem. 267, 20225–20232 (1992).
Wharton, S.A. et al. Electron microscopy of antibody complexes of influenza virus haemagglutinin in the fusion pH conformation. EMBO J. 14, 240–246 (1995).
Calder, L.J., Wasilewski, S., Berriman, J.A. & Rosenthal, P.B. Structural organization of a filamentous influenza A virus. Proc. Natl. Acad. Sci. USA 107, 10685–10690 (2010).
Carr, C.M. & Kim, P.S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73, 823–832 (1993).
Ivanovic, T., Choi, J.L., Whelan, S.P., van Oijen, A.M. & Harrison, S.C. Influenza-virus membrane fusion by cooperative fold-back of stochastically induced hemagglutinin intermediates. eLife 2, e00333 (2013).
Kim, Y.H. et al. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc. Natl. Acad. Sci. USA 108, 20992–20997 (2011).
Fontana, J., Cardone, G., Heymann, J.B., Winkler, D.C. & Steven, A.C. Structural changes in Influenza virus at low pH characterized by cryo-electron tomography. J. Virol. 86, 2919–2929 (2012).
Fontana, J. & Steven, A.C. At low pH, influenza virus matrix protein M1 undergoes a conformational change prior to dissociating from the membrane. J. Virol. 87, 5621–5628 (2013).
Harris, A. et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA 103, 19123–19127 (2006).
Lee, K.K. Architecture of a nascent viral fusion pore. EMBO J. 29, 1299–1311 (2010).
Vijayakrishnan, S. et al. Cryotomography of budding influenza A virus reveals filaments with diverse morphologies that mostly do not bear a genome at their distal end. PLoS Pathog. 9, e1003413 (2013).
Chlanda, P. et al. Structural analysis of the roles of influenza a virus membrane-associated proteins in assembly and morphology. J. Virol. 89, 8957–8966 (2015).
Fournier, E. et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 40, 2197–2209 (2012).
Noda, T. et al. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439, 490–492 (2006).
Wasilewski, S., Calder, L.J., Grant, T. & Rosenthal, P.B. Distribution of surface glycoproteins on influenza A virus determined by electron cryotomography. Vaccine 30, 7368–7373 (2012).
Kanaseki, T., Kawasaki, K., Murata, M., Ikeuchi, Y. & Ohnishi, S. Structural features of membrane fusion between influenza virus and liposome as revealed by quick-freezing electron microscopy. J. Cell Biol. 137, 1041–1056 (1997).
Ruigrok, R.W., Calder, L.J. & Wharton, S.A. Electron microscopy of the influenza virus submembranal structure. Virology 173, 311–316 (1989).
Diao, J. et al. Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. eLife 1, e00109 (2012).
Hernandez, J.M. et al. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science 336, 1581–1584 (2012).
Chernomordik, L.V. & Kozlov, M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 15, 675–683 (2008).
Ford, M.G. et al. Curvature of clathrin-coated pits driven by epsin. Nature 419, 361–366 (2002).
Bose, S., Jardetzky, T.S. & Lamb, R.A. Timing is everything: fine-tuned molecular machines orchestrate paramyxovirus entry. Virology 479-480, 518–531 (2015).
Harrison, S.C. Viral membrane fusion. Virology 479-480, 498–507 (2015).
Bharat, T.A. et al. SNARE and regulatory proteins induce local membrane protrusions to prime docked vesicles for fast calcium-triggered fusion. EMBO Rep. 15, 308–314 (2014).
Söllner, T.H. Intracellular and viral membrane fusion: a uniting mechanism. Curr. Opin. Cell Biol. 16, 429–435 (2004).
Jackson, D.C. et al. Electron microscopic evidence for the association of M2 protein with the influenza virion. Arch. Virol. 118, 199–207 (1991).
Wharton, S.A., Belshe, R.B., Skehel, J.J. & Hay, A.J. Role of virion M2 protein in influenza virus uncoating: specific reduction in the rate of membrane fusion between virus and liposomes by amantadine. J. Gen. Virol. 75, 945–948 (1994).
Skehel, J.J. & Schild, G.C. The polypeptide composition of influenza A viruses. Virology 44, 396–408 (1971).
Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Mastronarde, D.N. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343–352 (1997).
Acknowledgements
We are grateful to S. Wharton for advice on experiments, M. Maiorca for assistance in building segmentation models of the membranes, and J. Skehel and J. Molloy for discussions. Research was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council (program code U117581334 to P.B.R.), and the Wellcome Trust.
Author information
Authors and Affiliations
Contributions
L.J.C. and P.B.R. designed experiments and wrote the manuscript. L.J.C. performed experiments and analyses.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 2D cryomicroscopy images of low-pH time course after trypsin treatment (pH 7; pH 5, 1 min; pH 5, 5 min; pH 5, 30 min).
Images are unfiltered.
(a) pH7 images show no contact between virus and liposomes, which are content-less, sometimes multi-lamellar and heterogeneous in size.
(b) pH5, 1 min. Viral particles retain capsular shape but are in contact with many liposomes.
(c) pH5, 5 mins. Viral particles are less ordered and contacting many liposomes. Some particles have fused with liposomes and viral contents can be observed in some liposomes.
(d) pH5, 30 mins. Large liposomes contain viral protein structures, similar to those seen in low pH virions. Glycoproteins are widely distributed around the membrane and contacts with other liposomes occur.
Supplementary Figure 2 Tomogram section showing virus interactions with liposomes at the pH 5, 1-min time point.
Images are slices of tomograms (projections of a 22 Å slab) showing the morphology of the viral particles with intact M1 matrix layer, NA patch, and closely packed RNP segments labeled.
Supplementary Figure 3 Gallery of 2D images of the HA extended intermediate.
Images in every second row are identical to row above but indicate membranes (red lines) bridged by extended HA intermediate (green lines). images are filtered as described in methods
Supplementary Figure 4 Gallery of 3D tomogram sections of the HA extended intermediate.
Images are slices of tomograms (projections of 22 Å slab) showing additional examples to those in Figure 5a. Images in every second row are identical to row above but indicate membranes (red lines) bridged by extended HA intermediate (green lines).
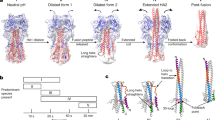
Supplementary Figure 5 Histograms of lengths of different HA structures observed and measured from tomograms.
(a) Length measurements for pH7 HA and HA extended intermediate. (b) Length measurements for bars, lines in star arrangements, and HA2. Model projections (right column) have been calculated from crystal structures described in Figure 1.
Supplementary Figure 6 Tomogram sections of a 3D contact zone between virus and liposome (1-min time point).
Images are tomogram slices (projections of a 13 Å slab) at intervals of 43 Å. Images in every second row are identical to row above but indicate membranes (red lines) bridged by extended HA intermediate (green lines). Last panel: shows membranes and extended HA intermediate for all tomogram sections. Corresponds to contact zones in Figure 4a-d.
Supplementary Figure 7 Tomogram sections of a 3D contact zone between virus and liposome (1-min time point).
Images are tomogram slices (projections of a 13 Å slab) at intervals of 43 Å. Images in every second row are identical to row above but indicating membranes (red lines) bridged by extended HA intermediate (green lines). Last panel: shows membranes and extended HA intermediate for all tomogram sections.
Supplementary Figure 8 Gallery of bar images.
Shows additional examples to those in Figure 5c,d of dense bars at membrane contacts (yellow arrows). First two panels of first row are tomogram sections. Remaining images are 2D images filtered as described in methods. Black spots densities are gold fiducial markers.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 3326 kb)
Rights and permissions
About this article
Cite this article
Calder, L., Rosenthal, P. Cryomicroscopy provides structural snapshots of influenza virus membrane fusion. Nat Struct Mol Biol 23, 853–858 (2016). https://doi.org/10.1038/nsmb.3271
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3271
This article is cited by
-
An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates
Virus Genes (2022)
-
In situ structure and organization of the influenza C virus surface glycoprotein
Nature Communications (2021)
-
The native structure of the assembled matrix protein 1 of influenza A virus
Nature (2020)
-
Structural proteomics, electron cryo-microscopy and structural modeling approaches in bacteria–human protein interactions
Medical Microbiology and Immunology (2020)