Abstract
The conserved oligomeric Golgi (COG) complex orchestrates vesicular trafficking to and within the Golgi apparatus. Here, we use negative-stain electron microscopy to elucidate the architecture of the hetero-octameric COG complex from Saccharomyces cerevisiae. Intact COG has an intricate shape, with four (or possibly five) flexible legs, that differs strikingly from that of the exocyst complex and appears to be well suited for vesicle capture and fusion.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Südhof, T.C. & Rothman, J.E. Science 323, 474–477 (2009).
Yu, I.M. & Hughson, F.M. Annu. Rev. Cell Dev. Biol. 26, 137–156 (2010).
Schindler, C., Chen, Y., Pu, J., Guo, X. & Bonifacino, J.S. Nat. Cell Biol. 17, 639–650 (2015).
Whyte, J.R. & Munro, S. J. Cell Sci. 115, 2627–2637 (2002).
Dong, G., Hutagalung, A.H., Fu, C., Novick, P. & Reinisch, K.M. Nat. Struct. Mol. Biol. 12, 1094–1100 (2005).
Pérez-Victoria, F.J. et al. Proc. Natl. Acad. Sci. USA 107, 12860–12865 (2010).
Richardson, B.C. et al. Proc. Natl. Acad. Sci. USA 106, 13329–13334 (2009).
Tripathi, A., Ren, Y., Jeffrey, P.D. & Hughson, F.M. Nat. Struct. Mol. Biol. 16, 114–123 (2009).
Vasan, N., Hutagalung, A., Novick, P. & Reinisch, K.M. Proc. Natl. Acad. Sci. USA 107, 14176–14181 (2010).
Ha, J.Y. et al. Proc. Natl. Acad. Sci. USA 111, 15762–15767 (2014).
Ren, Y. et al. Cell 139, 1119–1129 (2009).
Lees, J.A., Yip, C.K., Walz, T. & Hughson, F.M. Nat. Struct. Mol. Biol. 17, 1292–1297 (2010).
Freeze, H.H. & Ng, B.G. Cold Spring Harb. Perspect. Biol. 3, a005371 (2011).
Willett, R., Ungar, D. & Lupashin, V. Histochem. Cell Biol. 140, 271–283 (2013).
Miller, V.J. & Ungar, D. Traffic 13, 891–897 (2012).
Reynders, E. et al. Hum. Mol. Genet. 18, 3244–3256 (2009).
Ungar, D. et al. J. Cell Biol. 157, 405–415 (2002).
Heider, M.R. et al. Nat. Struct. Mol. Biol. 23, 59–66 (2016).
Lübbehusen, J. et al. Hum. Mol. Genet. 19, 3623–3633 (2010).
Foulquier, F. et al. Hum. Mol. Genet. 16, 717–730 (2007).
Ohi, M., Li, Y., Cheng, Y. & Walz, T. Biol. Proced. Online 6, 23–34 (2004).
Tang, G. et al. J. Struct. Biol. 157, 38–46 (2007).
Yang, Z., Fang, J., Chittuluru, J., Asturias, F.J. & Penczek, P.A. Structure 20, 237–247 (2012).
Acknowledgements
This work was supported by NIH grants R01 GM071574 (F.M.H.) and P01 GM062580 (T.W.).
Author information
Authors and Affiliations
Contributions
All authors designed experiments and analyzed data. J.Y.H., H.-T.C., D.U., and C.K.Y. performed the experiments. J.Y.H., H.-T.C., T.W., and F.M.H. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Purification and negative-stain EM of K. lactis Cog5–8.
(a) The purified recombinant K. lactis Cog5-8 complex, visualized by SDS-PAGE and Coomassie Blue staining, is shown together with a representative image field and ten representative class averages. (b) Complete gallery of K. lactis Cog5-8 class averages (side length of each panel = 38 nm). The 129 class averages, which account for 4,958 particles, were obtained by subjecting 8,825 particles to 14 generations of ISAC.
Supplementary Figure 2 Negative-stain EM of S. cerevisiae Cog1–8.
Complete gallery of S. cerevisiae Cog1-8 class averages (side length of each panel = 63 nm). The 278 class averages, which account for 5,182 particles, were obtained by subjecting 31,872 particles to 18 generations of ISAC.
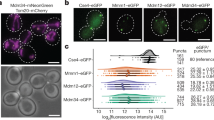
Supplementary Figure 3 Localizing individual subunits within the K. lactis Cog5–8 complex.
(a) Purified K. lactis Cog5-8 complexes containing single GFP tags, visualized by SDS-PAGE and Coomassie Blue staining. GFP-Cog6ΔN denotes N-terminally truncated Cog6 (residues 147-779). (b) Complete galleries of GFP-tagged K. lactis Cog5-8 class averages (side length of each panel = 38 nm). Only those complexes in which the GFP tag was visible in class averages are included; the remaining complexes were indistinguishable from the untagged complexes shown in Supplementary Fig. 1b.
Supplementary Figure 4 Dimensions of S. cerevisiae Cog1–8.
The indicated lengths (mean ± s.d.; n=5) were derived from class averages using ImageJ (https://imagej.nih.gov/ij/).
Supplementary Figure 5 Negative-stain EM of bovine COG.
Shown are four additional image fields of the bovine COG complex (see also Fig. 2e). The particles were too heterogeneous to allow successful class averaging.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Table 1 (PDF 1662 kb)
Supplementary Data Set 1
Uncropped gel for Fig. 1 (PDF 489 kb)
Yeast COG appears to contain flexible hinges
Negative-stain EM of S. cerevisiae Cog1–8 yields class averages in which the legs of the complex adopt various orientations. (MOV 163 kb)
Rights and permissions
About this article
Cite this article
Ha, J., Chou, HT., Ungar, D. et al. Molecular architecture of the complete COG tethering complex. Nat Struct Mol Biol 23, 758–760 (2016). https://doi.org/10.1038/nsmb.3263
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3263
This article is cited by
-
Crystal structure of Sec10, a subunit of the exocyst complex
Scientific Reports (2017)
-
CATCHR, HOPS and CORVET tethering complexes share a similar architecture
Nature Structural & Molecular Biology (2016)