Abstract
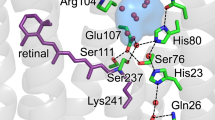
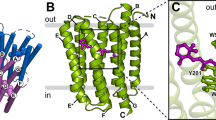
Conserved prolines in the transmembrane helices of G-protein-coupled receptors (GPCRs) are often considered to function as hinges that divide the helix into two segments capable of independent motion. Depending on their potential to hydrogen-bond, the free C=O groups associated with these prolines can facilitate conformational flexibility, conformational switching or stabilization of the receptor structure. To address the role of conserved prolines in family A GPCRs through solid-state NMR spectroscopy, we focus on bovine rhodopsin, a GPCR in the visual receptor subfamily. The free backbone C=O groups on helices H5 and H7 stabilize the inactive rhodopsin structure through hydrogen-bonds to residues on adjacent helices. In response to light-induced isomerization of the retinal chromophore, hydrogen-bonding interactions involving these C=O groups are released, thus facilitating repacking of H5 and H7 onto the transmembrane core of the receptor. These results provide insights into the multiple structural and functional roles of prolines in membrane proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sansom, M.S.P. & Weinstein, H. Hinges, swivels and switches: the role of prolines in signalling via transmembrane α-helices. Trends Pharmacol. Sci. 21, 445–451 (2000).
Cordes, F.S., Bright, J.N. & Sansom, M.S.P. Proline-induced distortions of transmembrane helices. J. Mol. Biol. 323, 951–960 (2002).
Williams, K.A. & Deber, C.M. Proline residues in transmembrane helices: structural or dynamic role? Biochemistry 30, 8919–8923 (1991).
von Heijne, G. Proline kinks in transmembrane α-helices. J. Mol. Biol. 218, 499–503 (1991).
Fu, Q. et al. Structural basis and functional role of intramembrane trimerization of the Fas/CD95 death receptor. Mol. Cell 61, 602–613 (2016).
Cao, Z. & Bowie, J.U. Shifting hydrogen bonds may produce flexible transmembrane helices. Proc. Natl. Acad. Sci. USA 109, 8121–8126 (2012).
Smith, S.O. Structure and activation of the visual pigment rhodopsin. Annu. Rev. Biophys. 39, 309–328 (2010).
Ballesteros, J.A. & Weinstein, H. Integrated methods for the construction of three dimensional models and computational probing of structure-function relations in G-protein coupled receptors. Meth. Neurosci. 25, 366–428 (1995).
Palczewski, K. et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745 (2000).
Li, J., Edwards, P.C., Burghammer, M., Villa, C. & Schertler, G.F.X. Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 343, 1409–1438 (2004).
Park, J.H., Scheerer, P., Hofmann, K.P., Choe, H.W. & Ernst, O.P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454, 183–187 (2008).
Deupi, X. et al. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc. Natl. Acad. Sci. USA 109, 119–124 (2012).
Choe, H.W. et al. Crystal structure of metarhodopsin II. Nature 471, 651–655 (2011).
Elling, C.E. et al. Metal ion site engineering indicates a global toggle switch model for seven-transmembrane receptor activation. J. Biol. Chem. 281, 17337–17346 (2006).
Schwartz, T.W., Frimurer, T.M., Holst, B., Rosenkilde, M.M. & Elling, C.E. Molecular mechanism of 7TM receptor activation: a global toggle switch model. Annu. Rev. Pharmacol. Toxicol. 46, 481–519 (2006).
Ganter, U.M., Gärtner, W. & Siebert, F. Rhodopsin-lumirhodopsin phototransition of bovine rhodopsin investigated by Fourier transform infrared difference spectroscopy. Biochemistry 27, 7480–7488 (1988).
Van Arnam, E.B., Lester, H.A. & Dougherty, D.A. Dissecting the functions of conserved prolines within transmembrane helices of the D2 dopamine receptor. ACS Chem. Biol. 6, 1063–1068 (2011).
Wess, J., Nanavati, S., Vogel, Z. & Maggio, R. Functional role of proline and tryptophan residues highly conserved among G protein-coupled receptors studied by mutational analysis of the m3 muscarinic receptor. EMBO J. 12, 331–338 (1993).
Stitham, J., Martin, K.A. & Hwa, J. The critical role of transmembrane prolines in human prostacyclin receptor activation. Mol. Pharmacol. 61, 1202–1210 (2002).
Imai, H. et al. Single amino acid residue as a functional determinant of rod and cone visual pigments. Proc. Natl. Acad. Sci. USA 94, 2322–2326 (1997).
Deupi, X. & Standfuss, J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr. Opin. Struct. Biol. 21, 541–551 (2011).
Deupi, X. Relevance of rhodopsin studies for GPCR activation. Biochim. Biophys. Acta 1837, 674–682 (2014).
Gullion, T. & Schaefer, J. Rotational-echo double-resonance NMR. J. Magn. Reson. 81, 196–200 (1989).
Saito, H. Conformation-dependent C-13 chemical-shifts: a new means of conformational characterization as obtained by high-resolution solid-state C-13 NMR. Magn. Reson. Chem. 24, 835–852 (1986).
Gu, Z.T., Zambrano, R. & McDermott, A. Hydrogen-bonding of carboxyl groups in solid state amino acids and peptides: comparison of carbon chemical shielding, infrared frequencies, and structures. J. Am. Chem. Soc. 116, 6368–6372 (1994).
Szilagyi, L. Chemical-shifts in proteins come of age. Prog. Nucl. Magn. Reson. Spectrosc. 27, 325–443 (1995).
Scheerer, P. et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 (2008).
Beck, M., Sakmar, T.P. & Siebert, F. Spectroscopic evidence for interaction between transmembrane helices 3 and 5 in rhodopsin. Biochemistry 37, 7630–7639 (1998).
Hamm, H.E. et al. Site of G protein binding to rhodopsin mapped with synthetic peptides from the alpha subunit. Science 241, 832–835 (1988).
White, S.H. & Wimley, W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 28, 319–365 (1999).
Rader, A.J. et al. Identification of core amino acids stabilizing rhodopsin. Proc. Natl. Acad. Sci. USA 101, 7246–7251 (2004).
Sung, C.H., Davenport, C.M. & Nathans, J. Rhodopsin mutations responsible for autosomal dominant retinitis pigmentosa: clustering of functional classes along the polypeptide chain. J. Biol. Chem. 268, 26645–26649 (1993).
Wigley, W.C. et al. A protein sequence that can encode native structure by disfavoring alternate conformations. Nat. Struct. Biol. 9, 381–388 (2002).
Goncalves, J.A. et al. Highly conserved tyrosine stabilizes the active state of rhodopsin. Proc. Natl. Acad. Sci. USA 107, 19861–19866 (2010).
Fritze, O. et al. Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc. Natl. Acad. Sci. USA 100, 2290–2295 (2003).
Isogai, S. et al. Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nature 530, 237–241 (2016).
Goncalves, J. et al. Magic angle spinning nuclear magnetic resonance spectroscopy of G protein-coupled receptors. Methods Enzymol. 522, 365–389 (2013).
Gullion, T. & Schaefer, J. in Advances in Magnetic Resonance Vol. 13 (ed. Warren, W.S.) 57–84 (Academic Press, 1989).
Eilers, M., Ying, W., Reeves, P.J., Khorana, H.G. & Smith, S.O. Magic angle spinning nuclear magnetic resonance of isotopically labeled rhodopsin. Methods Enzymol. 343, 212–222 (2002).
Jastrzebska, B., Goc, A., Golczak, M. & Palczewski, K. Phospholipids are needed for the proper formation, stability, and function of the photoactivated rhodopsin-transducin complex. Biochemistry 48, 5159–5170 (2009).
Farrens, D.L. & Khorana, H.G. Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J. Biol. Chem. 270, 5073–5076 (1995).
Acknowledgements
This work was supported by the National Institutes of Health (NIH) grant GM41412 (to S.O.S.) We thank H. Sasaki and X. Zhou (Institute of Protein Research, Osaka University) for expression and purification of several of the 15N-13C-labeled rhodopsin samples, and J. Goncalves for preliminary experiments with the Gα peptide. We thank J. Nathans (Johns Hopkins University) for providing the HEK293S cell line.
Author information
Authors and Affiliations
Contributions
M.E., P.J.R. and S.O.S. conceived the study; N.K., A.P. and M.E. prepared samples; M.E. and M.Z. collected and analyzed NMR data; A.P. and O.B.S.-R. analyzed the protein database for proline interactions; C.A.O. constructed rhodopsin mutants; and N.K., A.P., P.J.R. and S.O.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Notes 1 and 2 (PDF 924 kb)
Rights and permissions
About this article
Cite this article
Kimata, N., Pope, A., Sanchez-Reyes, O. et al. Free backbone carbonyls mediate rhodopsin activation. Nat Struct Mol Biol 23, 738–743 (2016). https://doi.org/10.1038/nsmb.3257
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3257
This article is cited by
-
Strategies for acquisition of resonance assignment spectra of highly dynamic membrane proteins: a GPCR case study
Journal of Biomolecular NMR (2023)
-
Retinal orientation and interactions in rhodopsin reveal a two-stage trigger mechanism for activation
Nature Communications (2016)