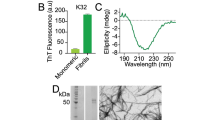
Abstract
Misfolded α-synuclein amyloid fibrils are the principal components of Lewy bodies and neurites, hallmarks of Parkinson's disease (PD). We present a high-resolution structure of an α-synuclein fibril, in a form that induces robust pathology in primary neuronal culture, determined by solid-state NMR spectroscopy and validated by EM and X-ray fiber diffraction. Over 200 unique long-range distance restraints define a consensus structure with common amyloid features including parallel, in-register β-sheets and hydrophobic-core residues, and with substantial complexity arising from diverse structural features including an intermolecular salt bridge, a glutamine ladder, close backbone interactions involving small residues, and several steric zippers stabilizing a new orthogonal Greek-key topology. These characteristics contribute to the robust propagation of this fibril form, as supported by the structural similarity of early-onset-PD mutants. The structure provides a framework for understanding the interactions of α-synuclein with other proteins and small molecules, to aid in PD diagnosis and treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Spillantini, M.G. et al. α-synuclein in Lewy bodies. Nature 388, 839–840 (1997).
Luk, K.C. et al. Exogenous α-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc. Natl. Acad. Sci. USA 106, 20051–20056 (2009).
Volpicelli-Daley, L.A. et al. Exogenous α-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron 72, 57–71 (2011).
Desplats, P. et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of α-synuclein. Proc. Natl. Acad. Sci. USA 106, 13010–13015 (2009).
Luk, K.C. et al. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338, 949–953 (2012).
Peelaerts, W. et al. α-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature 522, 340–344 (2015).
Heise, H. et al. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci. USA 102, 15871–15876 (2005).
Comellas, G. et al. Structured regions of α-synuclein fibrils include the early-onset Parkinson's disease mutation sites. J. Mol. Biol. 411, 881–895 (2011).
Gath, J. et al. Solid-state NMR sequential assignments of α-synuclein. Biomol. NMR Assign. 6, 51–55 (2012).
Bousset, L. et al. Structural and functional characterization of two α-synuclein strains. Nat. Commun. 4, 2575 (2013).
Wasmer, C. et al. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science 319, 1523–1526 (2008).
Iwata, K. et al. 3D structure of amyloid protofilaments of β2-microglobulin fragment probed by solid-state NMR. Proc. Natl. Acad. Sci. USA 103, 18119–18124 (2006).
Petkova, A.T., Yau, W.-M. & Tycko, R. Experimental constraints on quaternary structure in Alzheimer's β-amyloid fibrils. Biochemistry 45, 498–512 (2006).
Lu, J.-X. et al. Molecular structure of β-amyloid fibrils in Alzheimer's disease brain tissue. Cell 154, 1257–1268 (2013).
Schütz, A.K. et al. Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew. Chem. Int. Ed. Engl. 54, 331–335 (2015).
Xiao, Y. et al. Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease. Nat. Struct. Mol. Biol. 22, 499–505 (2015).
Kloepper, K.D., Woods, W.S., Winter, K.A., George, J.M. & Rienstra, C.M. Preparation of α-synuclein fibrils for solid-state NMR: expression, purification, and incubation of wild-type and mutant forms. Protein Expr. Purif. 48, 112–117 (2006).
Kloepper, K.D., Hartman, K.L., Ladror, D.T. & Rienstra, C.M. Solid-state NMR spectroscopy reveals that water is nonessential to the core structure of α-synuclein fibrils. J. Phys. Chem. B 111, 13353–13356 (2007).
Rutherford, N.J. et al. Studies of lipopolysaccharide effects on the induction of α-synuclein pathology by exogenous fibrils in transgenic mice. Mol. Neurodegener. 10, 32 (2015).
Comellas, G. & Rienstra, C.M. Protein structure determination by magic-angle spinning solid-state NMR, and insights into the formation, structure, and stability of amyloid fibrils. Annu. Rev. Biophys. 42, 515–536 (2013).
Castellani, F. et al. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature 420, 98–102 (2002).
Takegoshi, K., Nakamura, S. & Terao, T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344, 631–637 (2001).
Lange, A., Luca, S. & Baldus, M. Structural constraints from proton-mediated rare-spin correlation spectroscopy in rotating solids. J. Am. Chem. Soc. 124, 9704–9705 (2002).
Nieuwkoop, A.J., Wylie, B.J., Franks, W.T., Shah, G.J. & Rienstra, C.M. Atomic resolution protein structure determination by three-dimensional transferred echo double resonance solid-state nuclear magnetic resonance spectroscopy. J. Chem. Phys. 131, 095101 (2009).
Franks, W.T. et al. Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR. Proc. Natl. Acad. Sci. USA 105, 4621–4626 (2008).
Shen, Y. & Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 (2013).
Schwieters, C.D., Kuszewski, J.J. & Clore, G.M. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 48, 47–62 (2006).
Hutchinson, E.G. & Thornton, J.M. The Greek key motif: extraction, classification and analysis. Protein Eng. 6, 233–245 (1993).
Giasson, B.I., Murray, I.V., Trojanowski, J.Q. & Lee, V.M. A hydrophobic stretch of 12 amino acid residues in the middle of α-synuclein is essential for filament assembly. J. Biol. Chem. 276, 2380–2386 (2001).
Conway, K.A., Harper, J.D. & Lansbury, P.T. Accelerated in vitro fibril formation by a mutant α-synuclein linked to early-onset Parkinson disease. Nat. Med. 4, 1318–1320 (1998).
Khurana, R. et al. A general model for amyloid fibril assembly based on morphological studies using atomic force microscopy. Biophys. J. 85, 1135–1144 (2003).
Vilar, M. et al. The fold of α-synuclein fibrils. Proc. Natl. Acad. Sci. USA 105, 8637–8642 (2008).
Baxa, U. et al. Architecture of Ure2p prion filaments: the N-terminal domains form a central core fiber. J. Biol. Chem. 278, 43717–43727 (2003).
Wan, W. & Stubbs, G. Fiber diffraction of the prion-forming domain HET-s(218-289) shows dehydration-induced deformation of a complex amyloid structure. Biochemistry 53, 2366–2370 (2014).
Petkova, A.T. et al. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA 99, 16742–16747 (2002).
Atsmon-Raz, Y. & Miller, Y. A proposed atomic structure of the self-assembly of the non-amyloid-β-component of human α-synuclein as derived by computational tools. J. Phys. Chem. B 119, 10005–10015 (2015).
Rodriguez, J.A. et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature 525, 486–490 (2015).
Wan, W. & Stubbs, G. Fungal prion HET-s as a model for structural complexity and self-propagation in prions. Proc. Natl. Acad. Sci. USA 111, 5201–5206 (2014).
Ulmer, T.S., Bax, A., Cole, N.B. & Nussbaum, R.L. Structure and dynamics of micelle-bound human α-synuclein. J. Biol. Chem. 280, 9595–9603 (2005).
Lokappa, S.B. & Ulmer, T.S. α-synuclein populates both elongated and broken helix states on small unilamellar vesicles. J. Biol. Chem. 286, 21450–21457 (2011).
Penel, S., Morrison, R.G., Dobson, P.D., Mortishire-Smith, R.J. & Doig, A.J. Length preferences and periodicity in β-strands: antiparallel edge β-sheets are more likely to finish in non-hydrogen bonded rings. Protein Eng. 16, 957–961 (2003).
Crowther, R.A., Daniel, S.E. & Goedert, M. Characterisation of isolated α-synuclein filaments from substantia nigra of Parkinson's disease brain. Neurosci. Lett. 292, 128–130 (2000).
Gath, J. et al. Unlike twins: an NMR comparison of two α-synuclein polymorphs featuring different toxicity. PLoS One 9, e90659 (2014).
Polymeropoulos, M.H. et al. Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science 276, 2045–2047 (1997).
Krüger, R. et al. Ala30Pro mutation in the gene encoding α-synuclein in Parkinson's disease. Nat. Genet. 18, 106–108 (1998).
Zarranz, J.J. et al. The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 55, 164–173 (2004).
Appel-Cresswell, S. et al. α-synuclein p.H50Q, a novel pathogenic mutation for Parkinson's disease. Mov. Disord. 28, 811–813 (2013).
Lesage, S. et al. G51D α-synuclein mutation causes a novel parkinsonian-pyramidal syndrome. Ann. Neurol. 73, 459–471 (2013).
Lemkau, L.R. et al. Mutant protein A30P α-synuclein adopts wild-type fibril structure, despite slower fibrillation kinetics. J. Biol. Chem. 287, 11526–11532 (2012).
Lemkau, L.R. et al. Site-specific perturbations of α-synuclein fibril structure by the Parkinson's disease associated mutations A53T and E46K. PLoS One 8, e49750 (2013).
Comellas, G., Lemkau, L.R., Zhou, D.H., George, J.M. & Rienstra, C.M. Structural intermediates during α-synuclein fibrillogenesis on phospholipid vesicles. J. Am. Chem. Soc. 134, 5090–5099 (2012).
Lv, G. et al. Structural comparison of mouse and human α-synuclein amyloid fibrils by solid-state NMR. J. Mol. Biol. 420, 99–111 (2012).
Jiang, L. et al. Structure-based discovery of fiber-binding compounds that reduce the cytotoxicity of amyloid beta. eLife 2, e00857 (2013).
Bagchi, D.P. et al. Binding of the radioligand SIL23 to α-synuclein fibrils in Parkinson disease brain tissue establishes feasibility and screening approaches for developing a Parkinson disease imaging agent. PLoS One 8, e55031 (2013).
Guo, J.L. et al. Distinct α-synuclein strains differentially promote tau inclusions in neurons. Cell 154, 103–117 (2013).
Marley, J., Lu, M. & Bracken, C. A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR 20, 71–75 (2001).
Lemaster, D.M. & Kushlan, D.M. Dynamical mapping of E. coli thioredoxin via 13C NMR relaxation analysis. J. Am. Chem. Soc. 118, 9255–9264 (1996).
Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Comellas, G., Lopez, J.J., Nieuwkoop, A.J., Lemkau, L.R. & Rienstra, C.M. Straightforward, effective calibration of SPINAL-64 decoupling results in the enhancement of sensitivity and resolution of biomolecular solid-state NMR. J. Magn. Reson. 209, 131–135 (2011).
Morcombe, C.R. & Zilm, K.W. Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 162, 479–486 (2003).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Kuszewski, J., Gronenborn, A.M. & Clore, G.M. Improving the packing and accuracy of NMR structures with a pseudopotential for the radius of gyration. J. Am. Chem. Soc. 121, 2337–2338 (1999).
Grishaev, A. & Bax, A. An empirical backbone-backbone hydrogen-bonding potential in proteins and its applications to NMR structure refinement and validation. J. Am. Chem. Soc. 126, 7281–7292 (2004).
Bermejo, G.A., Clore, G.M. & Schwieters, C.D. Smooth statistical torsion angle potential derived from a large conformational database via adaptive kernel density estimation improves the quality of NMR protein structures. Protein Sci. 21, 1824–1836 (2012).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Murray, I.V.J. et al. Role of α-synuclein carboxy-terminus on fibril formation in vitro. Biochemistry 42, 8530–8540 (2003).
Wall, J.S. & Simon, M.N. Scanning transmission electron microscopy of DNA-protein complexes. Methods Mol. Biol. 148, 589–601 (2001).
Sen, A. et al. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked β-solenoid model of HET-s prion fibrils. J. Biol. Chem. 282, 5545–5550 (2007).
McDonald, M., Kendall, A., Tanaka, M., Weissman, J.S. & Stubbs, G. Enclosed chambers for humidity control and sample containment in fiber diffraction. J. Appl. Crystallogr. 41, 206–209 (2008).
Effenberger, H., Mereiter, K. & Zemann, J. Crystal structure refinements of magnesite, calcite, rhodochrosite, siderite, smithonite, and dolomite, with discussion of some aspects of the stereochemistry of calcite type carbonates. Z. Kristallogr. Cryst. Mater. 156, 233–243 (1981).
Fraser, R.D.B., Macrae, T.P., Miller, A. & Rowlands, R.J. Digital processing of fibre diffraction patterns. J. Appl. Crystallogr. 9, 81–94 (1976).
Bian, W., Wang, H., McCullough, I. & Stubbs, G. WCEN: a computer program for initial processing of fiber diffraction patterns. J. Appl. Crystallogr. 39, 752–756 (2006).
Chandrasekaran, R. & Stubbs, G. in International Tables for Crystallography 2nd edn., Vol. F (eds. Arnold, E., Himmel, D.M. & Rossman, M.G.) 444–450 (Wiley, 2012).
Holmes, K.C. & Leigh, J.B. The effect of disorientation on the intensity distribution of non-crystalline fibres. I. Theory. Acta Crystallogr. A 30, 635–638 (1974).
Acknowledgements
This study was supported by the US National Institutes of Health (NIH) (grants R01-GM073770 to C.M.R., P50-NS053488 to V.M.Y.L. and P01-AG002132 to G.S.) and used SSNMR instrumentation procured with the support of grant S10-RR025037 (to C.M.R.) from the NIH National Center for Research Resources (NCRR). M.D.T., A.J.N. and A.M.B. were supported as members of the NIH Molecular Biophysics Training Grant at the University of Illinois at Urbana-Champaign (T32-GM008276), and D.J.C. is supported by grant T32-AG000255. J.M.C. was supported by a US National Science Foundation Graduate Research Fellowship. C.D.S. is supported by the Intramural Research Program of the Center for Information Technology at NIH. The authors thank J. Wall and B. Lin (Brookhaven National Laboratory) for STEM MPL sample preparation and data collection. The Brookhaven National Laboratory STEM was an NIH-supported Resource Center (P41-EB2181), and additional support was provided by the US Department of Energy (DOE), Office of Biological and Environmental Research. TEM images were collected at the Frederick Seitz Materials Research Laboratory Central Facilities, University of Illinois, which are partially supported by the DOE under grants DE-FG02-07ER46453 and DE-FG02-07ER46471. The Voyager-DE STR MALDI TOF mass spectrometer was purchased in part with a grant from the NIH NCRR (S10-RR011966). The Stanford Synchrotron Radiation Lightsource (SSRL) is a national user facility operated by Stanford University on behalf of the DOE, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the DOE and the NIH. The authors thank M. Tang (College of Staten Island) for helpful discussions.
Author information
Authors and Affiliations
Contributions
M.D.T. analyzed and collected SSNMR data, performed structure calculations, analyzed structural features and was the primary author on the manuscript. G.C. analyzed and collected SSNMR data, performed MPL data analysis and provided input on the manuscript. A.J.N. analyzed and collected SSNMR data, performed initial structure calculations and provided input on the manuscript. D.J.C. performed the immunofluorescence, biochemical and biophysical assays and helped prepare the manuscript. D.A.B. prepared isotopically labeled samples and provided input on the manuscript. K.D.K. aided in sample preparation and manuscript preparation. J.M.C. created scripts for data conversion, performed analysis of final structures and provided input on the manuscript. J.K.K. contributed to the sample preparation methods. A.M.B. contributed to the mass spectrometry and solution NMR data and analysis. A.K. and W.W. prepared fiber diffraction samples and collected and analyzed fiber diffraction data. G.S. analyzed fiber diffraction data and aided in manuscript preparation. C.D.S. supported the development of structure calculations in XPLOR-NIH. V.M.Y.L., J.M.G. and C.M.R. were the primary investigators, designed the experiments and aided in manuscript preparation, data collection and interpretation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Mass spectrometry and solution NMR data for α-syn samples used in this study.
(a) Matrix-assisted laser-desorption and ionization time-of-flight (MALDI-TOF) mass spectrum of uniformly-13C,15N-enriched α-syn monomer (blue) and natural abundance monomer (red). The molecular mass of natural abundance α-syn monomer was calculated to be 14.46 kDa, in agreement with the experimental mass spectrum. The mass of 15.24 kDa for the U-13C,15N-labeled sample corresponded to a ~98% incorporation of 13C and 15N during protein expression. (b) 15N-1H HSQC spectrum of monomeric α-syn prior to fibrillization. The chemical shifts agreed well with the published assignments (Eliezer, D., Kutluay, E., Bussell, R. & Browne, G. Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 307, 1061–1073 (2001).)
Supplementary Figure 2 SSNMR spectroscopy data for α-syn fibrils, exhibiting cross-peaks indicative of long-range restraints and intermolecular registry of the fibrils.
(a) 2D 13C-13C SSNMR {1H}-13C-{1H-1H}-13C spectrum (Lange, A., Luca, S. & Baldus, M. Structural constraints from proton-mediated rare-spin correlation spectroscopy in rotating solids. J. Am. Chem. Soc. 124, 9704–9705 (2002).) of dilute α-syn fibrils (sample D from Table 1) showing unambiguous, long-range intramolecular correlations. Data were acquired at 750 MHz 1H frequency, 12.5 kHz MAS and 400 μs 1H-1H mixing time, with 12 days signal averaging. Red labels correspond to unambiguous long-range distances, including those from A78 to V48, from A69 to G93, from F94 to I88, and from V82 to A89. These intramolecular long-range restraints were inconsistent with structural models that exhibit a domain swap. (b-e) 2D 15N,13C SSNMR transferred echo double resonance (TEDOR, Jaroniec, C.P., Filip, C. & Griffin, R.G. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly 13C, 15N-labeled solids. J. Am. Chem. Soc. 124, 10728–10742 (2002).) data showing intermolecular interactions. The data were collected for four samples: (b) 1,3-13C glycerol,15N-labeled fibrils (sample B), signal averaged for 28 hr; (c) 50:50 [1,3-13C]glycerol, natural abundance (n.a.) nitrogen: n.a. carbon, 15N labeled (sample E), signal averaged for 81 hr; (d) [2-13C]glycerol,15N labeled (sample C), signal averaged for 113 hr; and (e) 50:50 2-13C glycerol, n.a. nitrogen: n.a. carbon, 15N labeled (sample F), signal averaged for 124 hr. Data was collected with mixing times of (c, e) 14.4 ms at 500 MHz 1H frequency, 11.1 kHz MAS, and (b, d) 16.0 ms at 600 MHz 1H frequency, 10.0 kHz MAS. Cross-peaks present in both parts b and c, or d and e, demonstrate a parallel, in-register arrangement (Debelouchina, G.T., Platt, G.W., Bayro, M.J., Radford, S.E. & Griffin, R.G. Intermolecular alignment in β2-microglobulin amyloid fibrils. J. Am. Chem. Soc. 132, 17077–17079 (2010).) (f–k) Spectra of sample B collected at 750 MHz 1H frequency, 12.5 kHz MAS (f-i) 2D 13C-13C spectra using dipolar-assisted rotational resonance (DARR, Takegoshi, K., Nakamura, S. & Terao, T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344, 631–637 (2001).) mixing times of (f) 50 ms, signal averaged for 10.9 hr, (g) 100 ms, signal averaged for 5.7 hr, (h) 200 ms, signal averaged for 12.0 hr, and (i) 300 ms, signal averaged for 12.7 hr. Several long-range correlations (red) were observed between the F94 aromatic ring resonances (CD and CE) and I88, A90 and A91. (j-k) 2D planes from a 3D 15N-13CO-13CX correlation spectrum signal averaged for 24 hr. Long-range correlations observed at this relatively short mixing time indicate internuclear distances <5 Å.
Supplementary Figure 3 Full-length structure of the α-syn fibril, illustrating important features in expanded regions.
(a) View along the fibril axis showing the highly ordered core and the disordered tails. (b) Side view showing the β-sheet packing between each monomer as well as the disordered tails. (c-f) Core interactions illustrating NMR distance restraints with dotted lines. (g-l) Backbone traces for neighboring monomers drawn in blue, yellow, orange and red. (g, h) Side and top views of the salt bridge from E46 to K80 of the neighboring monomer. (i, j) Side and top views of the I88-A91-F94 pocket perpendicular to the fibril axis, exhibiting short intermolecular distances. (k, l) Side and top view of the Q79 side chain exhibiting a glutamine ladder, with intermolecular hydrogen bonds involving Nɛ2 and Oɛ1 moieties.
Supplementary Figure 4 SSNMR internal validation of structural restraints and dihedral angles for the α-syn fibril structure.
(a) The backbone trace is shown in grey, and black lines represent unambiguous distance restraints. Blue lines correspond to the shortest observed distance among the possible ambiguous assignments. The resulting structure is the lowest energy conformer consistent with all of the data. (b-c) Ramachandran probability maps of accepted dihedral angle regions (Lovell, S.C. et al. Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins 50, 437–450 (2003).) and plotted in Chimera (Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).) for residues 41–100. The only residue not in the accepted range is E57, which is part of an unstructured loop and lacks NMR restraints in the simulated annealing calculations. All glycine residues are within the accepted Ramachandran space.
Supplementary Figure 5 Cross-validation of the SSNMR structure of α-syn fibrils with electron microscopy and X-ray fiber diffraction.
(a) Chemical shift comparison of the fibril sample for solid-state NMR before and after washing with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer and sonication for electron microscopy. (b) Expansions of overlaid 2D 13C-13C spectra of the U-13C, 15N α-syn fibril as prepared for solid-state NMR (red) and fibrils washed with HEPES buffer as the samples for electron microscopy (blue). Spectra were acquired at 600 MHz 1H frequency, 13.3 kHz magic-angle spinning (MAS) and 50 ms dipolar-assisted rotational resonance (DARR, Takegoshi, K., Nakamura, S. & Terao, T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344, 631–637 (2001).). (c) Equatorial intensity plot of the calculated and experimental fiber diffraction pattern of the α-syn fibril. The solid line shows the experimental fiber diffraction pattern (equatorial intensity projected from Figure 4c) compared to the simulated pattern (dotted line) from the atomic coordinates of the structure presented in Figure 3. The simulated pattern recapitulates the main features of the experimental pattern including the fine features near 0.12 Å−1. The correlation coefficient between the two patterns is 0.77, in good agreement with previous amyloid fibril fiber diffraction comparisons.
Supplementary Figure 6 Comparison of the Cα chemical-shift perturbations for three early-onset Parkinson's disease–mutant forms of α-syn, relative to the wild-type chemical shifts.
(a–c) Chemical shift perturbation plots of the absolute deviation of the Cα shifts for A30P, E46K, and A53T previously published (Lemkau, L.R. et al. Mutant protein A30P α-synuclein adopts wild-type fibril structure, despite slower fibrillation kinetics. J. Biol. Chem. 287, 11526-11532 (2012); Lemkau, L.R. et al. Site-specific perturbations of α-synuclein fibril structure by the Parkinson's disease associated mutations A53T and E46K. PLoS One 8, e49750 (2013).) relative to the wild-type chemical shifts used in this work.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Table 1 (PDF 3466 kb)
Rights and permissions
About this article
Cite this article
Tuttle, M., Comellas, G., Nieuwkoop, A. et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat Struct Mol Biol 23, 409–415 (2016). https://doi.org/10.1038/nsmb.3194
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3194
This article is cited by
-
Intramolecular interaction kinetically regulates fibril formation by human and mouse α-synuclein
Scientific Reports (2023)
-
The Sherpa hypothesis: Phenotype-Preserving Disordered Proteins stabilize the phenotypes of neurons and oligodendrocytes
npj Systems Biology and Applications (2023)
-
Pharmacological characterization of the small molecule 03A10 as an inhibitor of α-synuclein aggregation for Parkinson’s disease treatment
Acta Pharmacologica Sinica (2023)
-
Molecular pathology of neurodegenerative diseases by cryo-EM of amyloids
Nature (2023)
-
Development of a hydroxyflavone-labelled 4554W peptide probe for monitoring αS aggregation
Scientific Reports (2023)