Abstract
Post-translational modification of proteins by ubiquitin (Ub) and Ub-like modifiers regulates DNA replication. We have previously shown that chromatin around replisomes is rich in SUMO and poor in Ub, whereas mature chromatin exhibits an opposite pattern. How this SUMO-rich, Ub-poor environment is maintained at sites of DNA replication in mammalian cells remains unexplored. Here we identify USP7 as a replisome-enriched SUMO deubiquitinase that is essential for DNA replication. By acting on SUMO and SUMOylated proteins, USP7 counteracts their ubiquitination. Inhibition or genetic deletion of USP7 leads to the accumulation of Ub on SUMOylated proteins, which are displaced away from replisomes. Our findings provide a model explaining the differential accumulation of SUMO and Ub at replication forks and identify an essential role of USP7 in DNA replication that should be considered in the development of USP7 inhibitors as anticancer agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Lehmann, A.R. Ubiquitin-family modifications in the replication of DNA damage. FEBS Lett. 585, 2772–2779 (2011).
Zhang, W., Qin, Z., Zhang, X. & Xiao, W. Roles of sequential ubiquitination of PCNA in DNA-damage tolerance. FEBS Lett. 585, 2786–2794 (2011).
Havens, C.G. & Walter, J.C. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 25, 1568–1582 (2011).
Huang, T.T. et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8, 339–347 (2006).
Cohn, M.A. et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell 28, 786–797 (2007).
Nijman, S.M. et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 17, 331–339 (2005).
Sriramachandran, A.M. & Dohmen, R.J. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 1843, 75–85 (2014).
Galanty, Y., Belotserkovskaya, R., Coates, J. & Jackson, S.P. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26, 1179–1195 (2012).
Ragland, R.L. et al. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 27, 2259–2273 (2013).
Yin, Y. et al. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26, 1196–1208 (2012).
Ulrich, H.D. Two-way communications between ubiquitin-like modifiers and DNA. Nat. Struct. Mol. Biol. 21, 317–324 (2014).
Davis, E.J. et al. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat. Struct. Mol. Biol. 19, 1093–1100 (2012).
Mosbech, A. et al. DVC1 (C1orf124) is a DNA damage–targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat. Struct. Mol. Biol. 19, 1084–1092 (2012).
Gibbs-Seymour, I. et al. Ubiquitin-SUMO circuitry controls activated fanconi anemia ID complex dosage in response to DNA damage. Mol. Cell 57, 150–164 (2015).
Hendriks, I.A., Schimmel, J., Eifler, K., Olsen, J.V. & Vertegaal, A.C. Ubiquitin-specific protease 11 (USP11) deubiquitinates hybrid small ubiquitin-like modifier (SUMO)-ubiquitin chains to counteract RING finger protein 4 (RNF4). J. Biol. Chem. 290, 15526–15537 (2015).
Psakhye, I. & Jentsch, S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820 (2012).
Bursomanno, S. et al. Proteome-wide analysis of SUMO2 targets in response to pathological DNA replication stress in human cells. DNA Repair (Amst.) 25, 84–96 (2015).
Bursomanno, S., McGouran, J.F., Kessler, B.M., Hickson, I.D. & Liu, Y. Regulation of SUMO2 target proteins by the proteasome in human cells exposed to replication stress. J. Proteome Res. 14, 1687–1699 (2015).
Schimmel, J. et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol. Cell 53, 1053–1066 (2014).
Tammsalu, T. et al. Proteome-wide identification of SUMO2 modification sites. Sci. Signal. 7, rs2 (2014).
Xiao, Z. et al. System-wide analysis of SUMOylation dynamics in response to replication stress reveals novel small ubiquitin-like modified target proteins and acceptor lysines relevant for genome stability. Mol. Cell. Proteomics 14, 1419–1434 (2015).
Sirbu, B.M. et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25, 1320–1327 (2011).
Lopez-Contreras, A.J. et al. A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep. 3, 1105–1116 (2013).
Dungrawala, H. et al. The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell 59, 998–1010 (2015).
Li, M., Brooks, C.L., Kon, N. & Gu, W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 13, 879–886 (2004).
Li, M. et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416, 648–653 (2002).
Altun, M. et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 18, 1401–1412 (2011).
Chauhan, D. et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 22, 345–358 (2012).
Reverdy, C. et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem. Biol. 19, 467–477 (2012).
Kon, N. et al. Inactivation of HAUSP in vivo modulates p53 function. Oncogene 29, 1270–1279 (2010).
Alonso-de Vega, I., Martín, Y. & Smits, V.A. USP7 controls Chk1 protein stability by direct deubiquitination. Cell Cycle 13, 3921–3926 (2014).
Colleran, A. et al. Deubiquitination of NF-κB by ubiquitin-specific protease-7 promotes transcription. Proc. Natl. Acad. Sci. USA 110, 618–623 (2013).
de Bie, P., Zaaroor-Regev, D. & Ciechanover, A. Regulation of the Polycomb protein RING1B ubiquitination by USP7. Biochem. Biophys. Res. Commun. 400, 389–395 (2010).
Faustrup, H., Bekker-Jensen, S., Bartek, J., Lukas, J. & Mailand, N. USP7 counteracts SCFβTrCP- but not APCCdh1-mediated proteolysis of Claspin. J. Cell Biol. 184, 13–19 (2009).
Khoronenkova, S.V. & Dianov, G.L. USP7S-dependent inactivation of Mule regulates DNA damage signalling and repair. Nucleic Acids Res. 41, 1750–1756 (2013).
Lecona, E., Narendra, V. & Reinberg, D. USP7 cooperates with SCML2 to regulate the activity of PRC1. Mol. Cell. Biol. 35, 1157–1168 (2015).
Maertens, G.N., El Messaoudi-Aubert, S., Elderkin, S., Hiom, K. & Peters, G. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J. 29, 2553–2565 (2010).
Qian, J. et al. USP7 modulates UV-induced PCNA monoubiquitination by regulating DNA polymerase eta stability. Oncogene 34, 4791–4796 (2015).
Schwertman, P. et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat. Genet. 44, 598–602 (2012).
Song, M.S. et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP–PML network. Nature 455, 813–817 (2008).
van der Horst, A. et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 8, 1064–1073 (2006).
Zhang, P. et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat. Cell Biol. 16, 864–875 (2014).
Jagannathan, M. et al. A role for USP7 in DNA replication. Mol. Cell. Biol. 34, 132–145 (2014).
Du, Z. et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signal. 3, ra80 (2010).
Felle, M. et al. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 39, 8355–8365 (2011).
Qin, W., Leonhardt, H. & Spada, F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J. Cell. Biochem. 112, 439–444 (2011).
Sowa, M.E., Bennett, E.J., Gygi, S.P. & Harper, J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009).
Ge, X.Q., Jackson, D.A. & Blow, J.J. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 21, 3331–3341 (2007).
Ibarra, A., Schwob, E. & Méndez, J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. USA 105, 8956–8961 (2008).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Liang, Q. et al. A selective USP1–UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 10, 298–304 (2014).
Alabert, C. et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16, 281–293 (2014).
Tatham, M.H. et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 (2008).
Dantuma, N.P. & Hoppe, T. Growing sphere of influence: Cdc48/p97 orchestrates ubiquitin-dependent extraction from chromatin. Trends Cell Biol. 22, 483–491 (2012).
Magnaghi, P. et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat. Chem. Biol. 9, 548–556 (2013).
Uwada, J. et al. The p150 subunit of CAF-1 causes association of SUMO2/3 with the DNA replication foci. Biochem. Biophys. Res. Commun. 391, 407–413 (2010).
Weinstock, J. et al. Selective dual inhibitors of the cancer-related deubiquitylating proteases USP7 and USP47. ACS Med. Chem. Lett. 3, 789–792 (2012).
Toledo, L.I. et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat. Struct. Mol. Biol. 18, 721–727 (2011).
Lecona, E. et al. Upregulation of annexin A1 expression by butyrate in human colon adenocarcinoma cells: role of p53, NF-Y, and p38 mitogen-activated protein kinase. Mol. Cell. Biol. 28, 4665–4674 (2008).
Méndez, J. & Stillman, B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 (2000).
Ekholm-Reed, S. et al. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J. Cell Biol. 165, 789–800 (2004).
Lecona, E. et al. Polycomb protein SCML2 regulates the cell cycle by binding and modulating CDK/CYCLIN/p21 complexes. PLoS Biol. 11, e1001737 (2013).
López-Contreras, A.J., Gutierrez-Martinez, P., Specks, J., Rodrigo-Perez, S. & Fernandez-Capetillo, O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J. Exp. Med. 209, 455–461 (2012).
Gao, Z. et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 (2012).
Mourón, S. et al. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 20, 1383–1389 (2013).
Jackson, D.A. & Pombo, A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–1295 (1998).
Petermann, E., Orta, M.L., Issaeva, N., Schultz, N. & Helleday, T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37, 492–502 (2010).
Wis´niewski, J.R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Wis´niewski, J.R., Zougman, A. & Mann, M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8, 5674–5678 (2009).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Udeshi, N.D., Mertins, P., Svinkina, T. & Carr, S.A. Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc. 8, 1950–1960 (2013).
Acknowledgements
Research was funded by Fundación Botín, Banco Santander, through its Santander Universities Global Division, and by grants from the Spanish Ministry of Economy and Competitiveness (MINECO) (SAF2011-23753 and SAF2014-57791-REDC), Worldwide Cancer Research (12-0229), Fundació La Marato de TV3, the Howard Hughes Medical Institute and the European Research Council (ERC-617840) to O.F.-C.; by a Marie-Curie International Outgoing Fellowship (project no. 235705) from the Seventh Framework Programme for Research and Technological Development, Marie Curie Actions, and a grant from MINECO (BFU2014-55168-JIN) that was cofunded by European Regional Development Funds (FEDER) to E.L.; by grants from the Danish Council for Independent Research and the Danish National Research Foundation to A.J.L.-C.; and by a PhD fellowship from MINECO (BES-2012-05 2030) to J.S. We would like to thank W. Gu (Columbia University) for providing Usp7lox/lox MEFs, J. Chen (MD Anderson Cancer Center) for providing Rad18-deficient HCT116 cells and J. Alegre-Cebollada (Spanish National Center for Cardiovascular Research) for help in the use of PyMOL software.
Author information
Authors and Affiliations
Contributions
E.L. participated in most of the experiments of this study and wrote the paper. S.R.-A. and J. Mendez performed DNA fiber analyses. I.R. and J. Muñoz assisted with proteomics. J.S. and M.M. assisted with IF and HTM analyses. A.J.L.-C. assisted with iPOND experiments. O.F.-C. coordinated the study and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
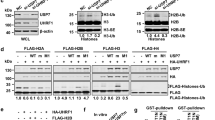
Supplementary Figure 1 USP7 inhibition stops DNA replication in a time- and dose-dependent manner.
(a-b) HCT116 cells were treated with increasing concentrations of P22077 or DMSO as a control for the indicated times. After incubation with 20 μM EdU for 30 min, cells were fixed and analyzed by HTM that quantified EdU (a) and γH2AX (b) levels per individual nucleus. DAPI was used to identify nuclei.
(c) Whole cell extracts of HCT116 cells treated with increasing concentrations of P22077 or DMSO (C) for the indicated times were analyzed by WB with antibodies against USP7, p21, p53 and MCM4.
Supplementary Figure 2 The effects of USP7 inhibition on DNA replication and replicative stress are independent of p53.
(a-b) Wild type (WT) and p53-deficient (p53 KO) MEF were treated with increasing concentrations of P22077 or DMSO (C) for the indicated times. After incubation with 20 μM EdU for 30 min, cells were fixed and analyzed by HTM that quantified EdU (a) and γH2AX (b) levels per individual nucleus.
(c) Representative IF examples illustrating the levels of EdU (green) and γH2AX (red) observed in WT and p53 KO MEF after 4 h of treatment. DAPI (blue) was used to identify nuclei. Scale bar, 20 μm.
Supplementary Figure 3 Two independent USP7 inhibitors recapitulate the effects of P22077, independently of PCNA ubiquitination.
(a-b) HCT116 cells were treated with increasing concentrations of P5091 (a) or HBX19818 (b), using DMSO as a control for the indicated times. After incubation with 20 μM EdU for 30 min, cells were fixed and analyzed by HTM that quantified EdU levels per individual nucleus. Equivalent results were obtained in two independent experiments.
(c-d) Whole cell extracts of HCT116 cells treated with increasing concentrations of P5091 or DMSO as a control for the indicated times were analyzed by WB with antibodies against USP7, PCNA, total and phosphorylated Chk1 (S345), and total and phosphorylated RPA2 (S4/S8) (c), or with antibodies against p53, p21 and MCM7 (d). Equivalent results were obtained in two independent experiments.
(e) Whole cell extracts of HCT116 wild-type (WT) and Rad18 KO cells treated with increasing concentrations of P22077 or DMSO (C) for the indicated times were analyzed by WB with antibodies against USP7, PCNA, total and phosphorylated Chk1 (S345), and total and phosphorylated RPA2 (S4/S8), and p21. Equivalent results were obtained in two independent experiments.
Supplementary Figure 4 Effect of USP7 overexpression on cell-cycle progression and origin firing.
(a) Cell cycle distribution of 293T-REx cells OneStrep-FLAG-HA-USP7 after being incubated for 1 day with (+Dox) or without (-Dox) 1 μg/ml doxycycline, measured by FACS staining with propidium iodide (PI) and BrdU. The percentage of BrdU cells is shown in (b).
(c) DNA fibers were extracted from 293T-REx OneStrep-FLAG-HA-USP7 cells incubated for 1 day with (+Dox) or without (-Dox) 1 μg/ml doxycycline and then sequentially treated with CldU and IdU. The firing of new origins was quantified as in Fig. 2e. ** p<0.01, t-test, two tails.
Supplementary Figure 5 Mass spectrometry analysis of the effect of USP7 on poly-SUMO2 and polyubiquitin chains.
(a) The intensity of the peptides for ubiquitin (black) and SUMO2 (grey) in the Ub-SUMO2X3 and the SUMO2X3 bands is shown.
(b) Di-Gly containing peptides for SUMO2 were identified and the cumulative intensity of all these peptides is shown for both bands.
(c) Poly-Ubiquitin (3-7) K48 or K63 chains were incubated with 25-100 nM USP7 or 100 nM USP1/UAF1 or in the absence of enzyme (C) for 2h at 37 °C. The products of the reaction were detected by Coomassie staining. The position of the different chains of Ubiquitin is indicated, together with the position of USP7 and the USP1/UAF1 dimmer.
(d) Poly-SUMO2 (3-8) chains were incubated with 20 nM USP7 or in the absence of enzyme (C) and the levels of SUMO2 were measured by WB.
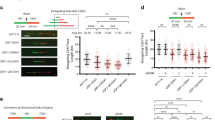
Supplementary Figure 6 Replication stress does not induce accumulation of SUMOylated proteins on chromatin.
(a) Representative IF images of U2OS cells treated with 50 μM P22077 or DMSO (C) for 4 h. Chromatin-bound levels of SUMO2 (red) were analyzed by a IF protocol that previously extracts the nuclear-soluble fraction of proteins. Besides this isolated example, the overall levels of chromatin-bound SUMO2/3 per individual nucleus as quantified by HTM are shown in Fig. 4e.
(b) HCT116 cells were treated with DMSO (C), 50 μM P22077 (P22) or 2-5 mM hydroxyurea (HU) for 2 h (left) or 4 h (right). The chromatin fraction was extracted and analyzed by Western blot with antibodies against SUMO2/3 or ubiquitin.
Supplementary Figure 7 Effects of inhibition of USP7 and/or p97 on DNA replication and effects of AdCre infection on WT MEFs.
(a-b) HCT116 cells were treated with 50 μM P22077 (P22), 5 μM NMS873 (NMS), both (P22-NMS) or DMSO as a control (C), for the indicated times. Chromatin fractions were analyzed by WB with antibodies against SUMO2/3 and USP7 (a), or Ub and histone H2A (b). Equivalent results were obtained in two independent experiments.
(c) HCT116 cells treated as in (a) were incubated with 20 μM EdU for 30 min. After incubation with 20 μM EdU for 30 min, cells were fixed and analyzed by HTM that quantified EdU levels per individual nucleus. The experiment was repeated three times and one representative experiment is shown.
(d) Western blot analysis of whole cell extracts from WT MEF mock infected (C) or infected with AdCre for 4 days. The levels of USP7 and CDK2 were measured using specific antibodies.
(e-f) DNA fibers were extracted 4 days after mock or AdCre infection of WT MEF and the fork rate (e) and percentage of origin firing (f) were measured. The experiments were repeated three times; the pool of the three experiments (fork rate) or the average (origin firing) is shown.
(g) Chromatin fraction of mock or AdCre infected WT MEF were assayed by Western blot with antibodies against SUMO2/3. Equivalent results were obtained in two independent experiments
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 to 1–7 (PDF 1088 kb)
Supplementary Data Set 1
Uncropped gel images (PDF 2760 kb)
Supplementary Table 1
iPOND data (XLS 370 kb)
Supplementary Table 2
Quantitative K-ɛ-GG site analysis for two HCT human cell biological replicates (XLSX 2390 kb)
Rights and permissions
About this article
Cite this article
Lecona, E., Rodriguez-Acebes, S., Specks, J. et al. USP7 is a SUMO deubiquitinase essential for DNA replication. Nat Struct Mol Biol 23, 270–277 (2016). https://doi.org/10.1038/nsmb.3185
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3185
This article is cited by
-
Concerted SUMO-targeted ubiquitin ligase activities of TOPORS and RNF4 are essential for stress management and cell proliferation
Nature Structural & Molecular Biology (2024)
-
The SUMOylation and ubiquitination crosstalk in cancer
Journal of Cancer Research and Clinical Oncology (2023)
-
DNA replication fork speed underlies cell fate changes and promotes reprogramming
Nature Genetics (2022)
-
Signalling mechanisms and cellular functions of SUMO
Nature Reviews Molecular Cell Biology (2022)
-
USP1-trapping lesions as a source of DNA replication stress and genomic instability
Nature Communications (2022)