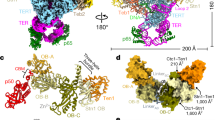
Abstract
Tetrahymena telomerase holoenzyme subunits p75, p45 and p19 form a subcomplex (7–4–1) peripheral to the catalytic core. We report structures of p45 and p19 and reveal them as the Stn1 and Ten1 subunits of the CST complex, which stimulates telomerase complementary-strand synthesis. 7–4–1 binds telomeric single-stranded DNA, and mutant p19 overexpression causes telomere 3′-overhang elongation. We propose that telomerase-tethered Tetrahymena CST coordinates telomere G-strand and C-strand synthesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Witkin, K.L. & Collins, K. Genes Dev. 18, 1107–1118 (2004).
Witkin, K.L., Prathapam, R. & Collins, K. Mol. Cell. Biol. 27, 2074–2083 (2007).
Min, B. & Collins, K. Mol. Cell 36, 609–619 (2009).
Zeng, Z. et al. Proc. Natl. Acad. Sci. USA 108, 20357–20361 (2011).
Upton, H.E., Hong, K. & Collins, K. Mol. Cell. Biol. 34, 4200–4212 (2014).
Jiang, J. et al. Nature 496, 187–192 (2013).
Hong, K. et al. Mol. Cell. Biol. 33, 3962–3971 (2013).
Sun, J. et al. Genes Dev. 23, 2900–2914 (2009).
Bryan, C., Rice, C., Harkisheimer, M., Schultz, D.C. & Skordalakes, E. PLoS ONE 8, e66756 (2013).
Gelinas, A.D. et al. Proc. Natl. Acad. Sci. USA 106, 19298–19303 (2009).
Stewart, J.A., Chaiken, M.F., Wang, F. & Price, C.M. Mutat. Res. 730, 12–19 (2012).
Chen, L.Y. & Lingner, J. Nucleus 4, 277–282 (2013).
Sun, J. et al. Cell Res. 21, 258–274 (2011).
Mer, G. et al. Cell 103, 449–456 (2000).
Jacob, N.K., Skopp, R. & Price, C.M. EMBO J. 20, 4299–4308 (2001).
Jacob, N.K., Kirk, K.E. & Price, C.M. Mol. Cell 11, 1021–1032 (2003).
Linger, B.R., Morin, G.B. & Price, C.M. Mol. Biol. Cell 22, 4161–4170 (2011).
Jacob, N.K., Lescasse, R., Linger, B.R. & Price, C.M. Mol. Cell. Biol. 27, 1592–1601 (2007).
Chen, L.Y., Redon, S. & Lingner, J. Nature 488, 540–544 (2012).
Ray, S., Karamysheva, Z., Wang, L., Shippen, D.E. & Price, C.M. Mol. Cell. Biol. 22, 5859–5868 (2002).
Otwinowski, Z. & Minor, W. Methods Enzymol. 276, 307–326 (1997).
Adams, P.D. et al. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Jerabek-Willemsen, M., Wienken, C.J., Braun, D., Baaske, P. & Duhr, S. Assay Drug Dev. Technol. 9, 342–353 (2011).
Moretti, P., Freeman, K., Coodly, L. & Shore, D. Genes Dev. 8, 2257–2269 (1994).
Shang, Y. et al. Proc. Natl. Acad. Sci. USA 99, 3734–3739 (2002).
Kimura, M. et al. Nat. Protoc. 5, 1596–1607 (2010).
Acknowledgements
We are grateful to the National Center for Protein Science Shanghai (Protein Expression and Purification system) for instrument support and technical assistance. We thank L. Wu, D. Yao, W. Qin and R. Zhang at Shanghai Synchrotron Radiation Facility for help with data collection and processing. This work was supported by grants from the Ministry of Science and Technology of China (2013CB910402 to M.L.), the National Natural Science Foundation of China (31330040 to M.L. and 31270787 to Z.Z.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08010201 to M.L.), the US National Science Foundation (Graduate Research Fellowship under grant DGE-1106400 to H.U.) and the US National Institutes of Health (GM54198 to K.C.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
B.W. and T.T. were responsible for the structural determination and biochemical experiments; H.U. for Tetrahymena telomerase activity assay and telomere-length quantification; J.S. for p45C crystallization; Y.Z., S.L., Z.Z. and J.C. for some of the protein purification; and J.S.B. for data collection and processing. K.C., J.W. and M.L. contributed to overall design, interpretation of the studies and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Superposition of p19 on the structures of human STN1, S. pombe Stn1 and S. cerevisiae Ten1.
(a) p19 vs. human STN1; (b) p19 vs. S. pombe Stn1; (c) p19 vs. S. cerevisiae Ten1. p19 is colored in cyan; HsSTN1, SpStn1, and ScTen1 are in light green, purple, and dark yellow, respectively. The rmsd values and the numbers of residues used for superposition are listed.
Supplementary Figure 2 Interaction of p19 and p45.
(a) Effects of p19 and p45 truncations on the p45-p19 interaction in yeast two-hybrid assays. Interaction between LexA-p19 and GAD-p45 was determined by measuring the β-galactosidase activity produced by the reporter gene. Data are averages of three independent β-galactosidase measurements. (b) and (c) Superposition of the p45N-p19 complex on the structures of human STN1N-TEN1 complex (b), and Candida tropicalis Stn1N-Ten1 complex (c). p45N and p19 are colored in yellow and cyan, respectively; HsSTN1N, HsTEN1, CtStn1N, and CtTen1 are shown in light purple, light orange, dark blue, and dark magenta, respectively. The rmsd values and the numbers of residues used for superposition are listed on the right.
Supplementary Figure 3 Superposition of p45C WH1 and WH2 motifs on the structures of S. cerevisiae and human STN1 WH motifs.
(a) p45C-WH1 vs. ScStn1C-WH1; (b) p45C-WH2 vs. ScStn1C-WH2; (c) p45C-WH1 vs. HsStn1C-WH1; (d) p45C-WH2 vs. HsStn1C-WH2. The rmsd values and the numbers of residues used for superposition are listed.
Supplementary Figure 4 Recombinant p75, p45 and p19 form a stable ternary complex in vitro.
(a) Gel filtration chromatography profile (Hiload Superdex 200) of the complex of full-length p75, p45, and p19 proteins coexpressed in Sf9 cells. (b) SDS-PAGE of p75, p45, and p19 proteins in the peak fractions of the gel filtration profile. Protein markers are indicated on the left. Elution volumes of the protein samples loaded in every lane in the gel are indicated on the top. (c) Effects of p75 and p45 truncations on the p75-p45 interaction in yeast two-hybrid assays. Interaction between LexA-p75 and GAD-p45 was determined by measuring the β-galactosidase activity produced by the reporter gene. Data are averages of three independent β-galactosidase measurements.
Supplementary Figure 5 Microscale thermophoresis measurements of the interactions between the 7–4–1 complex and wild-type and mutant telomeric ssDNAs.
A typical MST equilibrium experiment for quantification of an interaction is set up as a titration experiment. The concentration of a fluorescently labeled molecule is kept constant, whereas the concentration of an unlabeled binding partner is varied. This means that the target concentration is typically varied starting at a concentration of at least about 10 times above the expected dissociation constant down to sub-stoichiometric concentration with respect to the labeled molecule. The MST signal will detect the binding by a quantification of the change in the normalized fluorescence (i.e., the amplitude of the MST signal). By denoting x the fraction of labeled molecules bound to their targets, the changing fluorescence signal depending on the concentration of target is given by:
Fnorm = (1-x) Fnorm(unbound) + x Fnorm(bound)
Here, Fnorm (unbound) is the normalized fluorescence of unbound labeled molecules, and Fnorm (bound) is the normalized fluorescence of complexes (i.e., of labeled molecules bound to their targets in saturation). The unbound state acts as a reference state. All values are multiplied by a factor of 1,000, yielding a relative fluorescence change in per thousand [1/1000]. In our measurements, the 7-4-1 complex was titrated against 20 nM of the fluorescently labeled wild-type (a-c) or mutant (d-h) telomeric ssDNA. The calculated dissociation constant values of these DNAs to 7-4-1 are listed in each panel.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–3 (PDF 3916 kb)
Supplementary Data Set 1
Uncropped blots for Figure 2 (PDF 1487 kb)
Rights and permissions
About this article
Cite this article
Wan, B., Tang, T., Upton, H. et al. The Tetrahymena telomerase p75–p45–p19 subcomplex is a unique CST complex. Nat Struct Mol Biol 22, 1023–1026 (2015). https://doi.org/10.1038/nsmb.3126
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3126
This article is cited by
-
Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization
Nature Reviews Molecular Cell Biology (2021)
-
Structural insights into telomere protection and homeostasis regulation by yeast CST complex
Nature Structural & Molecular Biology (2020)
-
Tpz1TPP1 prevents telomerase activation and protects telomeres by modulating the Stn1-Ten1 complex in fission yeast
Communications Biology (2019)
-
Fine tuning the level of the Cdc13 telomere-capping protein for maximal chromosome stability performance
Current Genetics (2019)