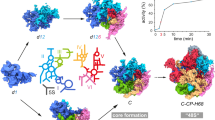
Abstract
Eukaryotic ribosome biogenesis involves a plethora of ribosome-assembly factors, and their temporal order of association with preribosomal RNA is largely unknown. By using Saccharomyces cerevisiae as a model organism, we developed a system that recapitulates and arrests ribosome assembly at early stages, thus providing in vivo snapshots of nascent preribosomal particles. Here we report the stage-specific order in which 70 ribosome-assembly factors associate with preribosomal RNA domains, thereby forming the 6-MDa small-subunit processome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Woolford, J.L. & Baserga, S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681 (2013).
Dragon, F. et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417, 967–970 (2002).
Osheim, Y.N. et al. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell 16, 943–954 (2004).
Phipps, K.R., Charette, J.M. & Baserga, S.J. The small subunit processome in ribosome biogenesis-progress and prospects. WIREs RNA 2, 1–21 (2011).
Turowski, T.W. & Tollervey, D. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip. Rev. RNA 6, 129–139 (2015).
Grandi, P. et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10, 105–115 (2002).
Pérez-Fernández, J., Román, A., De Las Rivas, J., Bustelo, X.R. & Dosil, M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 27, 5414–5429 (2007).
Marmier-Gourrier, N., Cléry, A., Schlotter, F., Senty-Ségault, V. & Branlant, C. A second base pair interaction between U3 small nucleolar RNA and the 5′-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Res. 39, 9731–9745 (2011).
Kudla, G., Granneman, S., Hahn, D., Beggs, J.D. & Tollervey, D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl. Acad. Sci. USA 108, 10010–10015 (2011).
Dutca, L.M., Gallagher, J.E.G. & Baserga, S.J. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 39, 5164–5180 (2011).
Granneman, S., Kudla, G., Petfalski, E. & Tollervey, D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. USA 106, 9613–9618 (2009).
Granneman, S., Petfalski, E., Swiatkowska, A. & Tollervey, D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 29, 2026–2036 (2010).
Lin, J., Lu, J., Feng, Y., Sun, M. & Ye, K. An RNA-binding complex involved in ribosome biogenesis contains a protein with homology to tRNA CCA-adding enzyme. PLoS Biol. 11, e1001669 (2013).
Turowski, T.W. et al. Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res. 42, 12189–12199 (2014).
Gupta, N. & Culver, G.M. Multiple in vivo pathways for Escherichia coli small ribosomal subunit assembly occur on one pre-rRNA. Nat. Struct. Mol. Biol. 21, 937–943 (2014).
Nogi, Y., Yano, R. & Nomura, M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. USA 88, 3962–3966 (1991).
Kirchhofer, A. et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–138 (2010).
Mitchell, P. Rrp47 and the function of the Sas10/C1D domain. Biochem. Soc. Trans. 38, 1088–1092 (2010).
Schuch, B. et al. The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. EMBO J. 33, 2829–2846 (2014).
Tackett, A.J. et al. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J. Proteome Res. 4, 1752–1756 (2005).
Soltanieh, S., Lapensée, M. & Dragon, F. Nucleolar proteins Bfr2 and Enp2 interact with DEAD-box RNA helicase Dbp4 in two different complexes. Nucleic Acids Res. 42, 3194–3206 (2014).
Granneman, S. et al. The nucleolar protein Esf2 interacts directly with the DExD/H box RNA helicase, Dbp8, to stimulate ATP hydrolysis. Nucleic Acids Res. 34, 3189–3199 (2006).
Wiederkehr, T., Prétôt, R.F. & Minvielle-Sebastia, L. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA 4, 1357–1372 (1998).
Lebaron, S. et al. Rrp5 binding at multiple sites coordinates pre-rRNA processing and assembly. Mol. Cell 52, 707–719 (2013).
Segerstolpe, Å. et al. Multiple RNA interactions position Mrd1 at the site of the small subunit pseudoknot within the 90S pre-ribosome. Nucleic Acids Res. 41, 1178–1190 (2012).
Thomson, E., Rappsilber, J. & Tollervey, D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA 13, 2165–2174 (2007).
Sardana, R. et al. The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PLoS Biol. 13, e1002083 (2015).
Ferreira-Cerca, S. et al. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat. Struct. Mol. Biol. 19, 1316–1323 (2012).
LeCuyer, K.A., Behlen, L.S. & Uhlenbeck, O.C. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry 34, 10600–10606 (1995).
Chernoff, Y.O., Vincent, A. & Liebman, S.W. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 13, 906–913 (1994).
Fridy, P.C. et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260 (2014).
Keefe, A.D., Wilson, D.S., Seelig, B. & Szostak, J.W. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr. Purif. 23, 440–446 (2001).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and Go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Acknowledgements
We thank Z. Hakhverdyan, J. Fernandez-Martinez and M. Rout for early advice with nanobody affinity purification, S. Gerstberger for technical assistance with northern blots and S. Liebman (University of Reno, Nevada) for the kind gift of yeast strains L-1491 and L-1496. M.C-M. is supported by a scholarship from Fonds de Recherche du Québec–Santé (FRQ-S). J.B. is supported by an European Molecular Biology Organization long-term fellowship (ALTF 51-2014). S.K. is supported by the Robertson Foundation, the Alfred P. Sloan Foundation, the Irma T. Hirschl Trust and the Human Frontier Science Program. The Proteomics Resource Center at Rockefeller University acknowledges funding from the Leona M. and Harry B. Helmsley Charitable Trust for mass spectrometer instrumentation.
Author information
Authors and Affiliations
Contributions
M.C.-M. and S.K. conceived the project. M.C.-M. performed all biochemical experiments. M.H. and J.B. established RNP protein-purification procedures. B.D.D. performed mass spectrometry analysis. M.C.-M. and S.K. analyzed the results and wrote the manuscript. All authors edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Simplified schematic of 35S pre-rRNA–processing steps in S. cerevisiae.
Processing intermediates of small subunit rRNA (18S, orange) and large subunit rRNAs (25S and 5.8S, blue and light blue respectively) of the 35S pre-rRNA are indicated. Key cleavage events are highlighted by descriptions in bold red with the responsible enzyme complexes listed in parentheses.
Supplementary Figure 2 Heat map of protein abundance as a function of transcript length for ribosome-assembly factors for which clear stage-specific assignments could be made.
Replicate 1 (identical sample as shown in Fig. 2b) and replicate 2 (a second independent experiment) are shown. Iog2 iBAQ values for relevant proteins identified in each construct were used to generate a heat-map of SSU processome assembly dynamics, where low abundance (<20) is shown in gray ranging to high abundance (>30) shown in red. Undetected signal is shown in black. Proteins are grouped into functional units, corresponding to the model presented in Figure 3. PolyA binding protein and streptavidin are shown at the top to represent internal controls. Proteins for which no clear stage-specific assignment could be made are listed in Supplementary Table 1 together with all original mass spectrometry data.
Supplementary Figure 3 A complex is formed between 26 proteins and the 5′ ETS in the presence of the MS2 tag, which does not inhibit ribosome assembly in vivo.
(a) SDS PAGE analysis of tandem affinity purified RNA-protein complexes with RNA and protein baits as indicated on the top. GFP-tagged UtpA (via Utp10), UtpB (via Utp1) and U3 snoRNP (via Rrp9) were used as baits to purify the 5´ ETS particle. Lanes 2 and 3 contain Utp10 with 3´ or 5´ tagged 5´ ETS sequences. Positions of protein baits, and proteins that are part of the purification scheme are indicated on the right. Corresponding mass spectrometry analysis is listed in Supplementary Table 1. (b) Northern blot analysis for 5´ MS2-tagged 5´ ETS transcripts shown in (a) using probes specific for the 5´ ETS (left) and the MS2 aptamer (right). Native 35S and 23S pre-rRNA intermediates are highlighted. (c) Growth phenotypes of yeast strains expressing a single copy of untagged (L-1496) or MS2-tagged (YSK145) rDNA. YPD plates were incubated for 1 day at 30 °C. (d) Schematic representations of the 35S pre-rRNA transcripts that are generated from pRDN4 (top) and pMS2x5-RDN4 (bottom). Boxes indicate positions of probes. (e) Northern blot analysis of pre-rRNAs from strains L-1496 and YSK145 using probes specific to the 5’ ETS (left) and the MS2 aptamer (right).
Supplementary Figure 4 Secondary structure and domains of S. cerevisiae 18S rRNA.
The diagram was modified from the Comparative RNA Web Site (http://www.rna.icmb.utexas.edu/) by using atomic resolution data for missing secondary structure annotations (pdb code 4V88). Domains are color-coded (5´ domain in green, central domain in yellow, 3´ major domain in orange and 3´ minor domain in red) with corresponding nucleotide numbers indicated. Nucleotide numbers are indicated in small font using the 18S sequence as reference, helices and expansion segments are indicated in large bold numbers. Base pairs involved in tertiary interactions are indicated with lines connecting circles or boxes around bases involved in the interactions. (-) corresponds to canonical base-pairs, while (•) and (°) correspond to non-canonical base-pairs.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 810 kb)
Supplementary Table 1
Mass spectrometry data (XLSX 246 kb)
Supplementary Table 2
Yeast strains (XLSX 23 kb)
Rights and permissions
About this article
Cite this article
Chaker-Margot, M., Hunziker, M., Barandun, J. et al. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol 22, 920–923 (2015). https://doi.org/10.1038/nsmb.3111
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3111
This article is cited by
-
Ribosome biogenesis gene DEF/UTP25 is essential for liver homeostasis and regeneration
Science China Life Sciences (2020)
-
Yeast R2TP Interacts with Extended Termini of Client Protein Nop58p
Scientific Reports (2019)
-
Ribosome assembly coming into focus
Nature Reviews Molecular Cell Biology (2019)
-
Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome
Nature Communications (2019)
-
Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate
Protein & Cell (2019)