Abstract
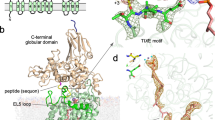
Sialyltransferases of the mammalian ST8Sia family catalyze oligo- and polysialylation of surface-localized glycoproteins and glycolipids through transfer of sialic acids from CMP–sialic acid to the nonreducing ends of sialic acid acceptors. The crystal structure of human ST8SiaIII at 1.85-Å resolution presented here is, to our knowledge, the first solved structure of a polysialyltransferase from any species, and it reveals a cluster of polysialyltransferase-specific structural motifs that collectively provide an extended electropositive surface groove for binding of oligo–polysialic acid chain products. The ternary complex of ST8SiaIII with a donor sugar analog and a sulfated glycan acceptor identified with a sialyltransferase glycan array provides insight into the residues involved in substrate binding, specificity and sialyl transfer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Varki, A. & Schauer, R. in Essentials of Glycobiology (Cold Spring Harbor Laboratory Press, 2009).
Sato, C. & Kitajima, K. Disialic, oligosialic and polysialic acids: distribution, functions and related disease. J. Biochem. 154, 115–136 (2013).
Crocker, P.R., Paulson, J.C. & Varki, A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 (2007).
Mühlenhoff, M., Rollenhagen, M., Werneburg, S., Gerardy-Schahn, R. & Hildebrandt, H. Polysialic acid: versatile modification of NCAM, SynCAM 1 and neuropilin-2. Neurochem. Res. 38, 1134–1143 (2013).
Acheson, A., Sunshine, J.L. & Rutishauser, U. NCAM polysialic acid can regulate both cell-cell and cell-substrate interactions. J. Cell Biol. 114, 143–153 (1991).
Schnaar, R.L., Gerardy-Schahn, R. & Hildebrandt, H. Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 94, 461–518 (2014).
Doherty, P., Cohen, J. & Walsh, F.S. Neurite outgrowth in response to transfected N-CAM changes during development and is modulated by polysialic acid. Neuron 5, 209–219 (1990).
Rutishauser, U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 9, 26–35 (2008).
Isomura, R., Kitajima, K. & Sato, C. Structural and functional impairments of polysialic acid by a mutated polysialyltransferase found in schizophrenia. J. Biol. Chem. 286, 21535–21545 (2011).
Falconer, R.A., Errington, R.J., Shnyder, S.D., Smith, P.J. & Patterson, L.H. Polysialyltransferase: a new target in metastatic cancer. Curr. Cancer Drug Targets 12, 925–939 (2012).
Cantarel, B.L. et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 (2009).
Angata, K. & Fukuda, M. Polysialyltransferases: major players in polysialic acid synthesis on the neural cell adhesion molecule. Biochimie 85, 195–206 (2003).
Foley, D.A., Swartzentruber, K.G. & Colley, K.J. Identification of sequences in the polysialyltransferases ST8Sia II and ST8Sia IV that are required for the protein-specific polysialylation of the neural cell adhesion molecule, NCAM. J. Biol. Chem. 284, 15505–15516 (2009).
Angata, K. et al. Differential biosynthesis of polysialic acid on neural cell adhesion molecule (NCAM) and oligosaccharide acceptors by three distinct alpha 2,8-sialyltransferases, ST8Sia IV (PST), ST8Sia II (STX), and ST8Sia III. J. Biol. Chem. 275, 18594–18601 (2000).
Yoshida, Y., Kojima, N., Kurosawa, N., Hamamoto, T. & Tsuji, S. Molecular cloning of Sia alpha 2,3Gal beta 1,4GlcNAc alpha 2,8-sialyltransferase from mouse brain. J. Biol. Chem. 270, 14628–14633 (1995).
Lee, Y.C. et al. Cloning and expression of cDNA for a human Sia alpha 2,3Gal beta 1, 4GlcNA:alpha 2,8-sialyltransferase (hST8Sia III). Arch. Biochem. Biophys. 360, 41–46 (1998).
Inoko, E. et al. Developmental stage-dependent expression of an alpha2,8-trisialic acid unit on glycoproteins in mouse brain. Glycobiology 20, 916–928 (2010).
Jarvis, D.L. Baculovirus-insect cell expression systems. Methods Enzymol. 463, 191–222 (2009).
Kojima, N., Tachida, Y., Yoshida, Y. & Tsuji, S. Characterization of mouse ST8Sia II (STX) as a neural cell adhesion molecule-specific polysialic acid synthase: requirement of core alpha1,6-linked fucose and a polypeptide chain for polysialylation. J. Biol. Chem. 271, 19457–19463 (1996).
Kitazume-Kawaguchi, S., Kabata, S. & Arita, M. Differential biosynthesis of polysialic or disialic acid structure by ST8Sia II and ST8Sia IV. J. Biol. Chem. 276, 15696–15703 (2001).
Blixt, O. et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA 101, 17033–17038 (2004).
Blixt, O. et al. Glycan microarrays for screening sialyltransferase specificities. Glycoconj. J. 25, 59–68 (2008).
Rao, F.V. et al. Structural insight into mammalian sialyltransferases. Nat. Struct. Mol. Biol. 16, 1186–1188 (2009).
Kuhn, B. et al. The structure of human alpha-2,6-sialyltransferase reveals the binding mode of complex glycans. Acta Crystallogr. D Biol. Crystallogr. 69, 1826–1838 (2013).
Meng, L. et al. Enzymatic basis for N-glycan sialylation: structure of rat alpha2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. J. Biol. Chem. 288, 34680–34698 (2013).
Datta, A.K. & Paulson, J.C. The sialyltransferase “sialylmotif” participates in binding the donor substrate CMP-NeuAc. J. Biol. Chem. 270, 1497–1500 (1995).
Geremia, R.A., Harduin-Lepers, A. & Delannoy, P. Identification of two novel conserved amino acid residues in eukaryotic sialyltransferases: implications for their mechanism of action. Glycobiology 7, v–vii (1997).
Jeanneau, C. et al. Structure-function analysis of the human sialyltransferase ST3Gal I: role of N-glycosylation and a novel conserved sialylmotif. J. Biol. Chem. 279, 13461–13468 (2004).
Wen, D.X. et al. Primary structure of Gal beta 1,3(4)GlcNAc alpha 2,3-sialyltransferase determined by mass spectrometry sequence analysis and molecular cloning: evidence for a protein motif in the sialyltransferase gene family. J. Biol. Chem. 267, 21011–21019 (1992).
Datta, A.K. Comparative sequence analysis in the sialyltransferase protein family: analysis of motifs. Curr. Drug Targets 10, 483–498 (2009).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Varki, A., Esko, J.D. & Colley, K.J. in Essentials of Glycobiology (Cold Spring Harbor Laboratory Press, 2009).
Angata, K., Yen, T.Y., El-Battari, A., Macher, B.A. & Fukuda, M. Unique disulfide bond structures found in ST8Sia IV polysialyltransferase are required for its activity. J. Biol. Chem. 276, 15369–15377 (2001).
Kulahin, N. et al. Direct demonstration of NCAM cis-dimerization and inhibitory effect of palmitoylation using the BRET2 technique. FEBS Lett. 585, 58–64 (2011).
Chiu, C.P. et al. Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat. Struct. Mol. Biol. 11, 163–170 (2004).
Qasba, P.K., Ramakrishnan, B. & Boeggeman, E. Substrate-induced conformational changes in glycosyltransferases. Trends Biochem. Sci. 30, 53–62 (2005).
Nakata, D., Zhang, L. & Troy, F.A. II. Molecular basis for polysialylation: a novel polybasic polysialyltransferase domain (PSTD) of 32 amino acids unique to the alpha 2,8-polysialyltransferases is essential for polysialylation. Glycoconj. J. 23, 423–436 (2006).
Audry, M. et al. Current trends in the structure-activity relationships of sialyltransferases. Glycobiology 21, 716–726 (2011).
Datta, A.K., Sinha, A. & Paulson, J.C. Mutation of the sialyltransferase S-sialylmotif alters the kinetics of the donor and acceptor substrates. J. Biol. Chem. 273, 9608–9614 (1998).
Klohs, W.D., Bernacki, R.J. & Korytnyk, W. Effects of nucleotides and nucleotide:analogs on human serum sialyltransferase. Cancer Res. 39, 1231–1238 (1979).
Lin, L.Y. et al. Structure and mechanism of the lipooligosaccharide sialyltransferase from Neisseria meningitidis. J. Biol. Chem. 286, 37237–37248 (2011).
Zielinska, D.F., Gnad, F., Wisniewski, J.R. & Mann, M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 (2010).
Funderburgh, J.L. Keratan sulfate: structure, biosynthesis, and function. Glycobiology 10, 951–958 (2000).
Tai, G.H., Huckerby, T.N. & Nieduszynski, I.A. Multiple non-reducing chain termini isolated from bovine corneal keratan sulfates. J. Biol. Chem. 271, 23535–23546 (1996).
Lauder, R.M., Huckerby, T.N. & Nieduszynski, I.A. Lectin affinity chromatography of articular cartilage fibromodulin: Some molecules have keratan sulphate chains exclusively capped by alpha(2–3)-linked sialic acid. Glycoconj. J. 28, 453–461 (2011).
Juneja, S.C. & Veillette, C. Defects in tendon, ligament, and enthesis in response to genetic alterations in key proteoglycans and glycoproteins: a review. Arthritis 2013, 154812 (2013).
Bentrop, J., Marx, M., Schattschneider, S., Rivera-Milla, E. & Bastmeyer, M. Molecular evolution and expression of zebrafish St8SiaIII, an alpha-2,8-sialyltransferase involved in myotome development. Dev. Dyn. 237, 808–818 (2008).
Angata, K., Chan, D., Thibault, J. & Fukuda, M. Molecular dissection of the ST8Sia IV polysialyltransferase: distinct domains are required for neural cell adhesion molecule recognition and polysialylation. J. Biol. Chem. 279, 25883–25890 (2004).
Zapater, J.L. & Colley, K.J. Sequences prior to conserved catalytic motifs of polysialyltransferase ST8Sia IV are required for substrate recognition. J. Biol. Chem. 287, 6441–6453 (2012).
Song, Y. et al. High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013).
Gray, J.J. et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 331, 281–299 (2003).
Foley, D.A., Swartzentruber, K.G., Lavie, A. & Colley, K.J. Structure and mutagenesis of neural cell adhesion molecule domains: evidence for flexibility in the placement of polysialic acid attachment sites. J. Biol. Chem. 285, 27360–27371 (2010).
Foley, D.A., Swartzentruber, K.G., Thompson, M.G., Mendiratta, S.S. & Colley, K.J. Sequences from the first fibronectin type III repeat of the neural cell adhesion molecule allow O-glycan polysialylation of an adhesion molecule chimera. J. Biol. Chem. 285, 35056–35067 (2010).
Mendiratta, S.S., Sekulic, N., Lavie, A. & Colley, K.J. Specific amino acids in the first fibronectin type III repeat of the neural cell adhesion molecule play a role in its recognition and polysialylation by the polysialyltransferase ST8Sia IV/PST. J. Biol. Chem. 280, 32340–32348 (2005).
Thompson, M.G., Foley, D.A. & Colley, K.J. The polysialyltransferases interact with sequences in two domains of the neural cell adhesion molecule to allow its polysialylation. J. Biol. Chem. 288, 7282–7293 (2013).
Close, B.E., Tao, K. & Colley, K.J. Polysialyltransferase-1 autopolysialylation is not requisite for polysialylation of neural cell adhesion molecule. J. Biol. Chem. 275, 4484–4491 (2000).
Mühlenhoff, M., Manegold, A., Windfuhr, M., Gotza, B. & Gerardy-Schahn, R. The impact of N-glycosylation on the functions of polysialyltransferases. J. Biol. Chem. 276, 34066–34073 (2001).
Baker, N.A., Sept, D., Joseph, S., Holst, M.J. & McCammon, J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98, 10037–10041 (2001).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Bunkóczi, G. & Read, R.J. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr. D Biol. Crystallogr. 67, 303–312 (2011).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Murshudov, G.N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Winn, M.D., Isupov, M.N. & Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 (2001).
Laskowski, R.A. PDBsum new things. Nucleic Acids Res. 37, D355–D359 (2009).
Pettersen, E.F. et al. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Gilbert, M. et al. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni: biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J. Biol. Chem. 277, 327–337 (2002).
Gosselin, S., Alhussaini, M., Streiff, M.B., Takabayashi, K. & Palcic, M.M. A continuous spectrophotometric assay for glycosyltransferases. Anal. Biochem. 220, 92–97 (1994).
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J.V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006).
Ishihama, Y., Rappsilber, J. & Mann, M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J. Proteome Res. 5, 988–994 (2006).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Cristobal, A. et al. In-house construction of a UHPLC system enabling the identification of over 4000 protein groups in a single analysis. Analyst 137, 3541–3548 (2012).
Scott, N.E. et al. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031-MCP201 (2011).
Neuhauser, N., Michalski, A., Cox, J. & Mann, M. Expert system for computer assisted annotation of MS/MS spectra. Mol. Cell. Proteomics 11, 1500–1509 (2012).
Schulz, B.L. & Aebi, M. Analysis of glycosylation site occupancy reveals a role for Ost3p and Ost6p in site-specific N-glycosylation efficiency. Mol. Cell. Proteomics 8, 357–364 (2009).
Eswar, N. et al. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 15, 5.6 (2006).
Acknowledgements
We wish to acknowledge the Consortium for Functional Glycomics (grants GM62116 and GM098791) for the sialyltransferase glycan array and for biotinylated CMP–Neu5Ac and the Canadian Light Source and Advanced Light Source for access to synchrotron data collection facilities. We also acknowledge operating funds from the Canadian Institutes of Health Research (N.C.J.S., S.G.W., W.W. and L.J.F.), the Howard Hughes Medical Institute International Senior Scholar program (N.C.J.S.) and the Canadian Foundation of Innovation and British Columbia Knowledge Development Fund (N.C.J.S., S.G.W. and L.J.F.). N.C.J.S. and S.G.W. are supported as Tier 1 Canada Research Chairs. G.V. is supported by fellowships from the Michael Smith Foundation for Health Research and the Canadian Department for Foreign Affairs and International Trade. L.B. is supported by the Deutsche Forschungsgemeinschaft. A Cystic Fibrosis Canada Fellowship supports E.L. The Ministry of Science and Technology, Taiwan (NSC-103-2917-I-564-009) supports C.-C.Y.
Author information
Authors and Affiliations
Contributions
G.V. designed the constructs and performed expression, purification, crystallization, data collection and processing as well as solution and refinement of crystallographic structures. L.J.W. phased the initial native structure, assisted with refinement of various data sets and performed Rosetta docking and modeling with protein acceptors. E.L. performed size-exclusion chromatography with multiangle light scattering experiments. G.A.W. performed biolayer interferometry experiments. L.B. and C.-C.Y. synthesized donor analog and acceptor compounds, and D.H.K. performed kinetic experiments. N.E.S. and L.J.F. performed and analyzed LC-MS experiments. W.W. contributed to analysis of glycan-array data. G.V., L.J.W., S.G.W. and N.C.J.S. designed the structural, mechanistic and kinetic experiments, analyzed the resulting data and wrote the manuscript. We thank A. Seitova (Structural Genomics Consortium Toronto) for reagents.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Purification and thin-layer chromatography (TLC) assays of ST8SiaIII.
(a) Elution profile of ST8SiaIII Δ60 examined by SEC-MALS with Coomassie-stained SDS-PAGE in the right top corner showing the crude expression medium of human ST8SiaIII Δ60 (50x concentrated, A), elution of the IMAC Ni2+ affinity column (B), elution of the S200 size-exclusion column (C), EndoH Hf (D) and PNGaseF (E) digested ST8SiaIII Δ60 and pure, concentrated ST8SiaIII Δ60 (F) protein. (b) TLC assay of wild type ST8SiaIII and mutant His354Ala with BODIPY-diSiaLac as acceptor, 1) no enzyme 2) positive control, ST8SiaIV Δ60 3) wild type ST8SiaIII Δ60 4) ST8SiaIII Δ60 mutant His354Ala. (c) TLC assay of wild type ST8SiaIII and ST8SiaII with BODIPY-diSiaLac as acceptor, 1) no enzyme 2) positive control 3) wild type ST8SiaIII Δ60 4) ST8SiaII Δ85. (d) TLC assay of wild type ST8SiaIII and mutants Asn190Ala, His337Ala with BODIPY-diSiaLac as acceptor, 1) no enzyme 2) positive control, ST8SiaIV Δ60 3) wild type ST8SiaIII Δ60 4) ST8SiaIII Δ60 mutant Asn190Ala 5) ST8SiaIII Δ60 mutant His337Ala.
Supplementary Figure 2 Topology of human ST8SiaIII.
(a) Topology diagram of human ST8SiaIII created with PDBsum (https://www.ebi.ac.uk/pdbsum/). (b) Overlap of monomer A of human ST8SiaIII (green) with human ST6GalI24 (4JS2, blue) bound to CMP (blue, ball & stick representation) and porcine ST3GalI23 (2WML, magenta) based on a secondary structure match (r.m.s.d. of 2.6 Å over ~200 common Cα). Note the conserved core of the sialyltransferases including the seven-stranded β-sheet around the donor-binding site.
Supplementary Figure 3 Sequence alignment of human ST8SiaIII with human ST8SiaII and ST8SiaIV.
Motifs highlighted (polybasic region, blue; large, green; polysialyltransferase domain, cyan; small, orange; III, black; very small, violet) and predicted transmembrane helix shown in grey. The header shows the most prevalent residues in small letters and conserved residues in capitalized red letters. Black asterisks refer to glycosylation sites Asn93, 113, 160 and 206 of ST8SiaIII. The alignment was created with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and UCSF Chimera68.
Supplementary Figure 4 Ternary-complex electron density maps, alternate conformations and comparison with porcine ST3GalI.
(a) Simulated annealing omit map of monomer A of the ternary complex (calculated with PHENIX in absence of donor and acceptor at 2.3 Å resolution; 2mFo-DFc (blue) contoured at 1σ, mFo-DFc (green) at 3σ, acceptor Sia-6S-LacNAc orange, donor analogue CMP-3FNeu5Ac dark green with disordered atom positions indicated at 85% transparency, His354 magenta. (b) Omit electron density of monomer B of the ternary complex (coloring see a). (c) Refined electron density map of monomer A (2mFo-DFc blue at 1σ, mFo-DFc green at 3σ). (d) Refined electron density map of monomer B (coloring see a). (e) Close-up on the simulated annealing omit map of the donor in monomer A of the ternary complex (coloring as in a). (f) Close-up on the simulated annealing omit map of the donor CMP-3FNeu5Ac in monomer B of the ternary complex (coloring as in a). (g) Overlap of Sia-6S-LacNAc of monomer A (yellow) on Sia-6S-LacNAc of monomer B (orange, r.m.s.d. 0.8 Å on all atoms) of the ternary complex. Note the different conformations of the glycerol group (black circle), with Sia-6S-LacNAc of monomer B exhibiting the catalytically permissive conformation for the C8 hydroxyl group. (h) Composite view of ternary complex of ST8SiaIII with CMP-3FNeu5Ac (monomer A) and Sia-6S-LacNAc (Sia-6S-LacNAc of monomer B, r.m.s.d of 0.8 Å to all atoms of Sia-6S-LacNAc of monomer A). (i) ST3GalI bound to acceptor and CMP23 (blue, PDB 2WNB).
Supplementary Figure 5 ESI-MS analysis and kinetics assay control to test for donor hydrolysis.
ESI-MS analysis of (a) axial CMP-3FNeu5Ac alone and (b) after 4 day incubation with ST8SiaIII Δ80. Data were recorded in the negative mode with dilutions ranging one order of magnitude. (c) CMP release (coupled to NADH oxidation measured by absorbance at 340 nm) using the assay conditions as described in methods, both before (in unfilled squares) and after (in filled circles) addition of ST8SiaIII to 1 μM. CMP-SA concentration was fixed at 1 mM and SiaLacNAc-6S concentration was varied. At 0 SiaLacNAc-6S, when the enzyme is added, there is no difference in the rate compared to the background control measurements before the enzyme is added. The background rate likely arises from spontaneous oxidation of NADH.
Supplementary Figure 6 Representative poses from top-scoring RosettaDock clusters in addition to the top pose shown in Figure 4d.
NCAM FN1 domain colored according to energy plot on Figure 4c. Superposed NCAM Ig5 domain (not used in docking) colored grey and distances between modified glycans Asn449 and Asn478 to active site Lys297 shown (diamond; only labeled in top left panel). The ST8SiaIV PBR is colored blue and the PSTD cyan. Residues implicated in the interaction are shown as spheres (NCAM FN1 acidic patch marked with a circle, ST8SiaIV PBR Arg93 marked with star, PBR Arg82 with a diamond; top left panel only) as is Asn74 (ST8SiaIV numbering), the major site of autopolysialylation (square; only labeled in top left panel).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Table 1 (PDF 6047 kb)
Supplementary Data Set 1
LC-MS/MS analysis of glycosylated ST8SiaIII (PDF 4657 kb)
Rights and permissions
About this article
Cite this article
Volkers, G., Worrall, L., Kwan, D. et al. Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation. Nat Struct Mol Biol 22, 627–635 (2015). https://doi.org/10.1038/nsmb.3060
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3060
This article is cited by
-
The vertebrate sialylation machinery: structure-function and molecular evolution of GT-29 sialyltransferases
Glycoconjugate Journal (2023)
-
Glycosyltransferases as targets for therapeutic intervention in cancer and inflammation: molecular modeling insights
Chemical Papers (2022)
-
Insights into the role of sialylation in cancer progression and metastasis
British Journal of Cancer (2021)
-
Synthetic O-acetylated sialosides facilitate functional receptor identification for human respiratory viruses
Nature Chemistry (2021)
-
Structural and functional role of disulphide bonds and substrate binding residues of the human beta-galactoside alpha-2,3-sialyltransferase 1 (hST3Gal1)
Scientific Reports (2019)