Abstract
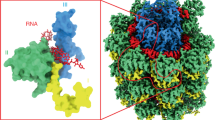
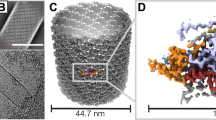
Flexible filamentous plant viruses cause more than half the viral crop damage in the world but are also potentially useful for biotechnology. Structural studies began more than 75 years ago but have failed, owing to the virion's extreme flexibility. We have used cryo-EM to generate an atomic model for bamboo mosaic virus, which reveals flexible N- and C-terminal extensions that allow deformation while still maintaining structural integrity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kendall, A. et al. J. Virol. 82, 9546–9554 (2008).
López-Moya, J. & García, J. in Encyclopedia of Virology Vol. 3, 1369–1375 (Academic Press, 1999).
Chen, T.H. et al. Virus Res. 166, 109–115 (2012).
Yang, C.D. et al. BMC Biotechnol. 7, 62 (2007).
Shukla, S. et al. Nanomedicine (Lond) 9, 221–235 (2014).
Zhang, T. et al. PLoS Biol. 4, e3 (2006).
Lico, C., Chen, Q. & Santi, L. J. Cell. Physiol. 216, 366–377 (2008).
Bernal, J.D. & Fankuchen, I. J. Gen. Physiol. 25, 147–165 (1941).
Beijerinck, M.W. Versl. Gew. Verg. Wis en Natuurk. Afd. 7, 229–235 (1898).
Namba, K. & Stubbs, G. Science 231, 1401–1406 (1986).
Ge, P. & Zhou, Z.H. Proc. Natl. Acad. Sci. USA 108, 9637–9642 (2011).
Richardson, J.F., Tollin, P. & Bancroft, J.B. Virology 112, 34–39 (1981).
Kendall, A. et al. Virology 436, 173–178 (2013).
Yang, S. et al. J. Mol. Biol. 422, 263–273 (2012).
Lin, N.S. et al. J. Gen. Virol. 75, 2513–2518 (1994).
Lin, M., Kitajima, E., Cupertino, F. & Costa, C. Phytopathology 67, 1439–1443 (1977).
Lan, P., Yeh, W.B., Tsai, C.W. & Lin, N.S. Mol. Plant Microbe Interact. 23, 903–914 (2010).
Egelman, E.H. eLife 3, e04969 (2014).
Leaver–Fay, A. et al. Methods Enzymol. 523, 109–143 (2013).
Hung, C.J. et al. Mol. Plant Pathol. 15, 196–210 (2014).
Chen, H.C. et al. Nucleic Acids Res. 40, 4641–4652 (2012).
Lin, N.-S. & Chen, C.-C. Phytopathology 81, 1551–1555 (1991).
Mindell, J.A. & Grigorieff, N. J. Struct. Biol. 142, 334–347 (2003).
Frank, J. et al. J. Struct. Biol. 116, 190–199 (1996).
Tang, G. et al. J. Struct. Biol. 157, 38–46 (2007).
Egelman, E.H. Ultramicroscopy 85, 225–234 (2000).
Rosenthal, P.B. & Henderson, R. J. Mol. Biol. 333, 721–745 (2003).
Song, Y. et al. Structure 21, 1735–1742 (2013).
DiMaio, F., Leaver–Fay, A., Bradley, P., Baker, D. & Andre, I. PLoS ONE 6, e20450 (2011).
DiMaio, F. et al. Nat. Methods 12, 361–365 (2015).
Acknowledgements
This work was supported by funds from US National Institutes of Health (NIH) GM035269, S10-RR025067 and S10-OD018149 (to E.H.E.); NSC98-2321-B-005-005-MY3 (to Y.-H.H.); NSC99-2628-B-001-012-MY3 and Academia Sinica (to N.-S.L.); and Scholarships for Excellent Students to Study Abroad from National Chung-Hsing University (to C.-C.C.). We thank K. Dryden for assistance with the cryo-EM. The cryo-EM work was conducted at the Molecular Electron Microscopy Core facility at the University of Virginia, which is supported by the School of Medicine and was built with NIH grant G20-RR31199. C.-C.C. thanks R.H. Cheng, L. Xing, R. Diaz, Z.H. Zhou and G.G. Liou for their assistance.
Author information
Authors and Affiliations
Contributions
C.-C.C. prepared the viral samples and did preliminary EM studies; Y.-H.H. and N.-S.L. initiated the studies, designed the viral constructs and provided guidance in the viral preparation; X.Y. did the EM and filament selection; E.H.E. did the image analysis and three-dimensional reconstruction; F.D. and B.F. did the molecular modeling; F.D. and E.H.E. wrote the paper; and all authors edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Power spectrum from BaMV segments.
A power spectrum generated from the 97,767 segments used in the final reconstruction (wt and Nd35 combined). The yellow arrow indicates the layer line (at 1/35.2 Å-1) arising from the left-handed one-start helix, while the red arrow indicates the fourth order of this one-start helical layer line at 1/8.8 Å-1. The log of the intensities is shown to allow for the large dynamic range.
Supplementary Figure 2 Map-map and model-map agreement.
(a) The FSC between the wildtype and Nd35 maps shows an apparent resolution of ~5.6 Å. (b) Models were fit to the wildtype maps (“training”) and evaluated against the Nd35 map (“testing”). The models show good agreement to both, with an FSC=0.5 crossing of about 5.0 Å, and with minimal overfitting (compare training map agreement to testing).
Supplementary Figure 3 Alternate views of the assembly, showing each subunit colored separately to better illustrate the intricate subunit interactions.
(a, b) Axial views of the assembly, looking from the top (a) and bottom (b) along the helical axis (referring to top and bottom from Figure 2). (c) The same view as in Figure 2c.
Supplementary Figure 4 Structure building and refinement are well converged.
(a) The best-scoring C-terminal models sampled by our backbone extension method show good convergence, with only small backbone deviations. (b,c) After all-atom refinement in the context of the full capsid, the best-scoring 5 models show minimal diversity in the core, with most of the diversity coming in the surface loops on the outside of the capsid (b), and the C-terminus on the inside of the capsid (c).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 641 kb)
Rights and permissions
About this article
Cite this article
DiMaio, F., Chen, CC., Yu, X. et al. The molecular basis for flexibility in the flexible filamentous plant viruses. Nat Struct Mol Biol 22, 642–644 (2015). https://doi.org/10.1038/nsmb.3054
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3054
This article is cited by
-
Atomic structure of potato virus X, the prototype of the Alphaflexiviridae family
Nature Chemical Biology (2020)
-
Origin of viruses: primordial replicators recruiting capsids from hosts
Nature Reviews Microbiology (2019)
-
Structure of Turnip mosaic virus and its viral-like particles
Scientific Reports (2019)
-
A versatile papaya mosaic virus (PapMV) vaccine platform based on sortase-mediated antigen coupling
Journal of Nanobiotechnology (2017)
-
RosettaES: a sampling strategy enabling automated interpretation of difficult cryo-EM maps
Nature Methods (2017)