Abstract
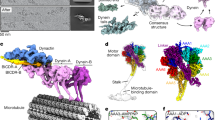
Cytoplasmic dynein associates with dynactin to drive cargo movement on microtubules, but the structure of the dynein–dynactin complex is unknown. Using electron microscopy, we determined the organization of native bovine dynein, dynactin and the dynein–dynactin–microtubule quaternary complex. In the microtubule-bound complex, the dynein motor domains are positioned for processive unidirectional movement, and the cargo-binding domains of both dynein and dynactin are accessible.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kardon, J.R. & Vale, R.D. Nat. Rev. Mol. Cell Biol. 10, 854–865 (2009).
Carter, A.P. J. Cell Sci. 126, 705–713 (2013).
Roberts, A.J., Kon, T., Knight, P.J., Sutoh, K. & Burgess, S.A. Nat. Rev. Mol. Cell Biol. 14, 713–726 (2013).
Bingham, J.B., King, S.J. & Schroer, T.A. Methods Enzymol. 298, 171–184 (1998).
Vallee, R.B., Wall, J.S., Paschal, B.M. & Shpetner, H.S. Nature 332, 561–563 (1988).
Amos, L.A. J. Cell Sci. 93, 19–28 (1989).
Steffen, W., Hodgkinson, J.L. & Wiche, G. J. Struct. Biol. 117, 227–235 (1996).
Vale, R.D. Cell 112, 467–480 (2003).
Wilkerson, C.G., King, S.M., Koutoulis, A., Pazour, G.J. & Witman, G.B. J. Cell Biol. 129, 169–178 (1995).
Hall, J., Song, Y., Karplus, P.A. & Barbar, E. J. Biol. Chem. 285, 22566–22575 (2010).
Urnavicius, L. et al. Science doi: 10.1126/science.aaa4080 (12 February 2015).
Tynan, S.H., Gee, M.A. & Vallee, R.B. J. Biol. Chem. 275, 32769–32774 (2000).
Schlager, M.A., Hoang, H.T., Urnavicius, L., Bullock, S.L. & Carter, A.P. EMBO J. 33, 1855–1868 (2014).
Burgess, S.A., Walker, M.L., Sakakibara, H., Oiwa, K. & Knight, P.J. J. Struct. Biol. 146, 205–216 (2004).
Schroer, T.A. Annu. Rev. Cell Dev. Biol. 20, 759–779 (2004).
Eckley, D.M. et al. J. Cell Biol. 147, 307–320 (1999).
Yeh, T.Y. et al. EMBO J. 32, 1023–1035 (2013).
Yeh, T.Y., Quintyne, N.J., Scipioni, B.R., Eckley, D.M. & Schroer, T.A. Mol. Biol. Cell 23, 3827–3837 (2012).
Hodgkinson, J.L., Peters, C., Kuznetsov, S.A. & Steffen, W. Proc. Natl. Acad. Sci. USA 102, 3667–3672 (2005).
Imai, H., Narita, A., Maeda, Y. & Schroer, T.A. J. Mol. Biol. 426, 3262–3271 (2014).
McKenney, R.J., Huynh, W., Tanenbaum, M.E., Bhabha, G. & Vale, R.D. Science 345, 337–341 (2014).
King, S.J. & Schroer, T.A. Nat. Cell Biol. 2, 20–24 (2000).
Bingham, J.B., King, S.J. & Schroer, T.A. Methods Enzymol. 298, 171–184 (1998).
Schroer, T.A. & Sheetz, M.P. J. Cell Biol. 115, 1309–1318 (1991).
Bertani, G. J. Bacteriol. 62, 293–300 (1951).
Suloway, C. et al. J. Struct. Biol. 151, 41–60 (2005).
Lander, G.C. et al. J. Struct. Biol. 166, 95–102 (2009).
Mindell, J.A. & Grigorieff, N. J. Struct. Biol. 142, 334–347 (2003).
Roseman, A.M. J. Struct. Biol. 145, 91–99 (2004).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. J. Struct. Biol. 128, 82–97 (1999).
Sorzano, C.O. et al. J. Struct. Biol. 148, 194–204 (2004).
Ogura, T., Iwasaki, K. & Sato, C. J. Struct. Biol. 143, 185–200 (2003).
Yang, Z., Fang, J., Chittuluru, J., Asturias, F.J. & Penczek, P.A. Structure 20, 237–247 (2012).
Tang, G. et al. J. Struct. Biol. 157, 38–46 (2007).
Hohn, M. et al. J. Struct. Biol. 157, 47–55 (2007).
Li, X. et al. Nat. Methods 10, 584–590 (2013).
Scheres, S.H. J. Struct. Biol. 180, 519–530 (2012).
Heymann, J.B. & Belnap, D.M. J. Struct. Biol. 157, 3–18 (2007).
Goddard, T.D., Huang, C.C. & Ferrin, T.E. J. Struct. Biol. 157, 281–287 (2007).
Kelley, L.A. & Sternberg, M.J. Nat. Protoc. 4, 363–371 (2009).
Screpanti, E. et al. Curr. Biol. 21, 391–398 (2011).
Acknowledgements
We thank R. McKenney (R. Vale laboratory, University of California, San Francisco) for providing the BicD2N construct, and the S. Encalada laboratory (Scripps) for providing mouse brains. We are enormously grateful to R. Milligan for his ongoing support and input into this project. This work was supported by the Damon Runyon Cancer Research Foundation (DFS-#07-13), the Pew Scholars program, the Searle Scholars program, and US National Institutes of Health (NIH) grant DP2 EB020402-01 to G.C.L. and NIH grant GM44589 to T.A.S.
Author information
Authors and Affiliations
Contributions
S.A.K. prepared the isolated dynein and dynactin. S.C. prepared the MT–DDB complex and performed all electron microscopy, image analysis and reconstructions. All authors contributed to the experimental design and assembly of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
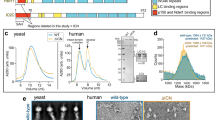
Supplementary Figure 1 Negative-stain EM of native cytoplasmic dynein.
(a) A representative micrograph of negatively stained dynein, with the flexible dimeric molecules circled in white. (b) Reference-free 2D class averages of dynein, showing the canonical “V” shaped structure with head domains spaced at varying distances.
Supplementary Figure 2 2D structural analysis of flexible dynein.
(a) Masking and assembly strategy used to generate detailed 2D averages of the entire dynein molecule. Reference-free 2D class averages of dynein are determined using traditional methods. An example of one of these averages is shown on the far left. Due to the flexibility of the link between the head and tail domains, the structural features of this region are blurred. Soft masks (dotted lines) are used to isolate each head and the tail region of the molecule, and the particles contributing to each class are subjected to 2D analysis (grey arrows on left), revealing finer structural features of the tail and head domains. Next, the mask is removed from the aligned particles and new unmasked class averages are generated, allowing visualization of surrounding structural features. The kinked region of the HC, which is seen in the unmasked class averages of both the head and tail domains, is then correlated to align the components to generate a complete dynein molecule (right image). (b) A montage of masked class averages for the tail and head regions (upper and lower rows, respectively).
Supplementary Figure 3 Negative-stain reconstruction of dynactin.
(a) A representative micrograph with individual particles circled in white, and reference-free 2D class averages on the right. (b) Class averages of dynactin with the flexible globular domain of p150Glued in a range of positions (indicated by yellow arrow). (c) On the left is a model of the 50 nm p150Glued coiled coil, with a filtered density representing the size of the CAP-Gly (PDB ID: 1TXQ). The globular domain in the class averages (right) is ∼20nm from the shoulder. The inset shows a focused class average of the globular domain, showing that the density is much larger than a CAP-Gly. (d) Surface rendering of the 24Å negative stain dynactin reconstruction shown from 4 different views (top panel), with corresponding forward projections of the reconstruction in the middle panel. Matching reference-free class averages are shown in the bottom panel. (e) FSC plot for the 3D reconstruction. The resolution using a 0.5 cutoff is ∼24Å. (f) Angular distribution of particles contributing to 3D reconstruction, with height of each cylinder related to the number of particles at that Euler angle.
Supplementary Figure 4 Cryo-EM reconstruction of dynactin.
(a) A representative cryo-electron micrograph of dynactin with individual particles denoted by white circles. (b) Reference-free 2D class averages showing the shoulder-less dynactin particles. (c) The α-helical “tentacles” of the CapZα and CapZβ subunits (orange and red, respectively) can be seen bound to the first Arp1 subunit of the short and long Arp strands, respectively. (d) Local resolution estimation of the 3D map shows a range of resolutions, from 6Å to 10 Å (shown by a spectrum of colors from blue to red). The Arp filament is the best-resolved component, and the pointed end complex is worst resolved, indicating increased flexibility in this region. (e) Gold Standard FSC plot for the 3D reconstruction of dynactin Arp filament, reporting a resolution of ∼6.5Å at a 0.143 cutoff. (f) Angular distribution of particles contributing to the final reconstruction.
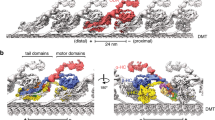
Supplementary Figure 5 Visualization of the native dynein–dynactin–BicD2 complex attached to the microtubule surface.
(a) AMP-PMP, but not ATP, causes dynein to bind MTs. Purified dynein (without added nucleotide) was supplemented with AMP-PNP, ATP or no nucleotide as shown and combined with phosphocellulose-purified, taxol-stabilized MTs. The MTs were pelleted through sucrose, then resusupended in buffer containing 10 mM ATP and pelleted again. The supernatant and pellet from the first centrifugation step (depleted supe and MT pellet) and the final supernatant (ATP release) were analyzed by SDS-PAGE. (b) SDS-Polyacrylamide gel electrophoresis (SDS-PAGE) of dynein-dynactin-BicD2N complex, isolated from mouse brain bound to microtubules. All the components of dynein and dynactin, as well as BicD2N and tubulin are present. (c) Negative stain micrograph showing DDB complexes (indicated by white arrow) attached to the MT surface. Although they are all pointed in the same direction, the DDB complexes are observed at a range of angles relative to the MT surface. (d) Several hundred DDB complexes at a range of angles relative to the MT surface were averaged together to determine the overall dimensions of the complex. The mean distance between the pointed tip of DDB from MT surface is ∼ 33nm, the center of head to the MT surface ∼17nm, and the length of the DDB molecule from the center of head to pointed tip is ∼43nm. (e) Focused classification (see Supplementary figure 2) was used to obtain detailed averages of the motor domains, showing that they are in close proximity to one another when attached to the MT surface. (f) Distribution of angles between DDB and MT surface. The majority of the DDB molecules are oriented between ∼20° and 60° relative to the MT surface. (g) Range of distances between the two heads in isolated dynein and DDB complexes, showing dramatically reduced inter-head distances in the incorporated dynein.
Supplementary information
Supplementary Figures
Supplementary Figures 1–5 (PDF 4197 kb)
Rights and permissions
About this article
Cite this article
Chowdhury, S., Ketcham, S., Schroer, T. et al. Structural organization of the dynein–dynactin complex bound to microtubules. Nat Struct Mol Biol 22, 345–347 (2015). https://doi.org/10.1038/nsmb.2996
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2996
This article is cited by
-
N-acetyl-D-glucosamine kinase binds dynein light chain roadblock 1 and promotes protein aggregate clearance
Cell Death & Disease (2020)
-
The regulatory function of the AAA4 ATPase domain of cytoplasmic dynein
Nature Communications (2020)
-
Dynactin1 depletion leads to neuromuscular synapse instability and functional abnormalities
Molecular Neurodegeneration (2019)
-
Cargo adaptors regulate stepping and force generation of mammalian dynein–dynactin
Nature Chemical Biology (2019)
-
Electron tomography of mouse LINC complexes at meiotic telomere attachment sites with and without microtubules
Communications Biology (2019)