Abstract
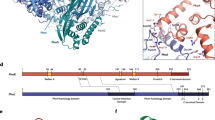
Bacteria use vitamin C (L-ascorbic acid) as a carbon source under anaerobic conditions. The phosphoenolpyruvate-dependent phosphotransferase system (PTS), comprising a transporter (UlaA), a IIB-like enzyme (UlaB) and a IIA-like enzyme (UlaC), is required for the anaerobic uptake of vitamin C and its phosphorylation to L-ascorbate 6-phosphate. Here, we present the crystal structures of vitamin C–bound UlaA from Escherichia coli in two conformations at 1.65-Å and 2.35-Å resolution. UlaA forms a homodimer and exhibits a new fold. Each UlaA protomer consists of 11 transmembrane segments arranged into a ‘V-motif’ domain and a ‘core’ domain. The V motifs form the interface between the two protomers, and the core-domain residues coordinate vitamin C. The alternating access of the substrate from the opposite side of the cell membrane may be achieved through rigid-body rotation of the core relative to the V motif.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Brown, L.A.S. & Jones, D.P. in Handbook of Antioxidants (eds. Cadenas, E. & Packer, L.) 117–156 (Marcel Dekker, New York, 1996).
Ellis, R.W. Vitamin-C deficiency and periostitis of both ulnae? Scurvy. Proc. R. Soc. Med. 32, 139–141 (1939).
Liang, W.-J., Johnson, D. & Jarvis, S.M. Vitamin C transport systems of mammalian cells. Mol. Membr. Biol. 18, 87–95 (2001).
Wang, H. et al. Human Na+-dependent vitamin C transporter 1 (hSVCT1): primary structure, functional characteristics and evidence for a non-functional splice variant. Biochim. Biophys. Acta 1461, 1–9 (1999).
Esselen, W.B. & Fuller, J.E. The oxidation of ascorbic acid as influenced by intestinal bacteria. J. Bacteriol. 37, 501–521 (1939).
Young, R.M. & James, L.H. Action of intestinal microorganisms on ascorbic acid. J. Bacteriol. 44, 75–84 (1942).
Zhang, Z., Aboulwafa, M., Smith, M.H. & Saier, M.H. Jr. The ascorbate transporter of Escherichia coli. J. Bacteriol. 185, 2243–2250 (2003).
Yew, W.S. & Gerlt, J.A. Utilization of l-ascorbate by Escherichia coli K-12: assignments of functions to products of the yjf-sga and yia-sgb operons. J. Bacteriol. 184, 302–306 (2002).
Postma, P.W., Lengeler, J.W. & Jacobson, G.R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57, 543–594 (1993).
Lengeler, J.W., Jahreis, K. & Wehmeier, U.F. Enzymes II of the phospho enol pyruvate-dependent phosphotransferase systems: their structure and function in carbohydrate transport. Biochim. Biophys. Acta 1188, 1–28 (1994).
Robillard, G.T. & Broos, J. Structure/function studies on the bacterial carbohydrate transporters, enzymes II, of the phosphoenolpyruvate-dependent phosphotransferase system. Biochim. Biophys. Acta 1422, 73–104 (1999).
Siebold, C., Flukiger, K., Beutler, R. & Erni, B. Carbohydrate transporters of the bacterial phosphoenolpyruvate: sugar phosphotransferase system (PTS). FEBS Lett. 504, 104–111 (2001).
Deutscher, J. et al. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78, 231–256 (2014).
Deutscher, J., Francke, C. & Postma, P.W. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031 (2006).
Saier, M.H., Hvorup, R.N. & Barabote, R.D. Evolution of the bacterial phosphotransferase system: from carriers and enzymes to group translocators. Biochem. Soc. Trans. 33, 220–224 (2005).
Chang, A.B., Lin, R., Studley, W.K., Tran, C.V. & Saier, M.H. Phylogeny as a guide to structure and function of membrane transport proteins. Mol. Membr. Biol. 21, 171–181 (2004).
Chen, J.S. et al. Phylogenetic characterization of transport protein superfamilies: superiority of SuperfamilyTree programs over those based on multiple alignments. J. Mol. Microbiol. Biotechnol. 21, 83–96 (2011).
Nguyen, T.X., Yen, M.R., Barabote, R.D. & Saier, M.H. Jr. Topological predictions for integral membrane permeases of the phosphoenolpyruvate:sugar phosphotransferase system. J. Mol. Microbiol. Biotechnol. 11, 345–360 (2006).
Cao, Y. et al. Crystal structure of a phosphorylation-coupled saccharide transporter. Nature 473, 50–54 (2011).
Hvorup, R., Chang, A.B. & Saier, M.H. Bioinformatic analyses of the bacterial l-ascorbate phosphotransferase system permease family. J. Mol. Microbiol. Biotechnol. 6, 191–205 (2003).
Linster, C.L., Van Schaftingen, E. & Vitamin, C. Biosynthesis, recycling and degradation in mammals. FEBS J. 274, 1–22 (2007).
Saier, M.H. Jr. & Reizer, J. The bacterial phosphotransferase system: new frontiers 30 years later. Mol. Microbiol. 13, 755–764 (1994).
Yernool, D., Boudker, O., Jin, Y. & Gouaux, E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 431, 811–818 (2004).
Crisman, T.J., Qu, S., Kanner, B.I. & Forrest, L.R. Inward-facing conformation of glutamate transporters as revealed by their inverted-topology structural repeats. Proc. Natl. Acad. Sci. USA 106, 20752–20757 (2009).
Von Heijne, G. & Gavel, Y. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174, 671–678 (1988).
Martinez-Jéhanne, V. et al. Role of the vpe carbohydrate permease in Escherichia coli urovirulence and fitness in vivo. Infect. Immun. 80, 2655–2666 (2012).
Ehrnstorfer, I.A., Geertsma, E.R., Pardon, E., Steyaert, J. & Dutzler, R. Crystal structure of a SLC11 (NRAMP) transporter reveals the basis for transition-metal ion transport. Nat. Struct. Mol. Biol. 21, 990–996 (2014).
Jaehme, M., Guskov, A. & Slotboom, D.J. Crystal structure of the vitamin B3 transporter PnuC, a full-length SWEET homolog. Nat. Struct. Mol. Biol. 21, 1013–1015 (2014).
Shi, Y. Common folds and transport mechanisms of secondary active transporters. Annu. Rev. Biophys. 42, 51–72 (2013).
Vinothkumar, K.R. & Henderson, R. Structures of membrane proteins. Q. Rev. Biophys. 43, 65–158 (2010).
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966).
Boudker, O. & Verdon, G. Structural perspectives on secondary active transporters. Trends Pharmacol. Sci. 31, 418–426 (2010).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data. Methods Enzymol. 276, 307–326 (1997).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 (1999).
Cowtan, K. dm: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 (1994).
Emsley, P., Lohkamp, B., Scott, W. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Vriend, G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8, 52–56 (1990).
Acknowledgements
We thank Shanghai Synchrotron Radiation Source for access to beamline BL17U, SPring-8 for access to beamline BL41XU and the Brookhaven National Synchrotron Light Source for access to beamline X29A. We thank N. Yan and E. Coutavas for discussion and comments on the manuscript and D. King (University of California, Berkeley) for MS analysis. This work was supported by funds from the Ministry of Science and Technology of China (grant nos. 2011CB911102 and 2015CB910104), Tsinghua University 985 Phase II funds, National Natural Science Foundation of China (31321062) and Beijing Municipal Commissions of Education and Science and Technology to J.W. We acknowledge the China National Center for Protein Sciences, Beijing, for providing the facility support. X.L. is supported as the Gordon and Betty Moore Foundation Fellow of the Life Sciences Research Foundation.
Author information
Authors and Affiliations
Contributions
P.L., X.Y., X.L. and J.W. designed all experiments. P.L., X.Y., W.W., S.F. and X.L. performed the experiments. All authors analyzed the data. P.L., X.Y., X.L. and J.W. contributed to manuscript preparation. J.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Sequence alignment of E. coli UlaA with homologs from other species and organisms and E. coli UlaA topology diagram.
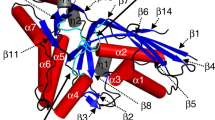
(a) Secondary structural elements of UlaA are indicated above the sequence alignment. Conserved amino acids are colored red and yellow in decreasing degrees of conservation, and those only conserved in TM11 of various UlaAs, not including VpeC, are colored green. UlaA is spatially organized into “V motif 1” (cyan), “Core 1” (yellow), “V motif 2” (Magenta), “Core 2” (orange), and “TM11” (gray) subdomains. The listed UlaA homologs include UlaA from Escherichia coli (GI: 606808039), Corynebacterium pseudotuberculosis (GI: 503005968), Streptomyces gancidicus (GI: 493093377), Bacillus licheniformis (GI: 489271278), Vibrio cholerae (GI: 669371971), Pasteurella multocida (GI: 504480874), Mycoplasma pulmonis (GI: 499227462), and VpeC from Escherichia coli (GI: 446267492). In the bacteria, e.g. Vibrio cholera, Pasteurella multocida, and Mycoplasma pulmonis, UlaB domains are fused C-terminal to the UlaA domains. The sequences were aligned with ClustalW (Thompson, J.D., Gibson, T. & Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Current protocols in bioinformatics, 2.3. 1-2.3. 22 (2002)). (b) The helices of UlaA are denoted as cylinders and β-sheet as arrows. The diagram is oriented with the extracellular side on top. The helix coloring scheme is consistent with that used in Fig. 1a. The black lines show the approximate location of the membrane, and the N- and C-termini are labelled. The structural repeats are highlighted by the trapezoids, which emphasize their relative orientation. The TM segments comprising the transporter cores are denoted as “Core 1” and “Core 2”, while those comprising the V motif are denoted as “V motif 1” and “V motif 2”, respectively, as in Fig. 1c.
Supplementary Figure 2 Representative electron density map of UlaA.
The stereo view of the 2Fo-Fc electron density of UlaA in C2 crystal form is shown. The electron density is contoured at 1.5 σ.
Supplementary Figure 3 Electrostatic-potential surface representations of UlaA dimer and superimposition of the two ‘inverted’ structural repeats within UlaA.
(a) The structure of the UlaA dimer is shown in cartoon and electrostatic potential surface representations as viewed within the plane of the membrane, represented as a grey rectangle. UlaA is spatially organized into “V motif 1” (cyan), “Core1” (yellow), “V motif 2” (Magenta), “Core 2” (orange), and “TM11” (gray) subdomains. Vitamin C is shown in ball-and-stick representation. (b) Viewed from the intracellular side of the membrane. (c) Superimposition of “Core 1” and “Core 2” subdomains. (d) Superimposition of “V motif 1” and “V motif 2” subdomains.
Supplementary Figure 4 L-ascorbate coordination by UlaA.
(a) Vitamin C is located in a concave pocket. The “Core 1” and “Core 2” subdomains of UlaA are shown in surface electrostatic potential. (b) The 2Fo-Fc electron density for vitamin C after refinement is contoured at 1.5 σ.
Supplementary Figure 5 Measurement of dissociation constants between UlaA variants and vitamin C by isothermal titration calorimetry (ITC).
Raw data are shown in the upper panels and the integrated heats per injection are plotted in the lower panel. The solid lines through the data are fits to the single site model. (a) Wild-type data. (e) The ligand (6 mM vitamin C) is titrated into buffer as a control. ITC data for UlaA variants with hydrogen-bond contributions are shown in (b-d) and (f-i). Data with van der Waals contributions are shown in (j-l). All the ITC experiments were performed independently three times.
Supplementary Figure 6 Structural changes between two crystal forms.
(a) Structural differences between the outward-open state and occluded state, shown from the periplasm, highlighting the overall rigid-body rotation of the core domain relative to the V motif domain. The disorder region in P21B is indicated by black ellipse. (b) Distances of Cα atoms between the core domains of C2A' and P21B molecules. The color scheme used is the same as that in Fig. 1c.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 4828 kb)
Rights and permissions
About this article
Cite this article
Luo, P., Yu, X., Wang, W. et al. Crystal structure of a phosphorylation-coupled vitamin C transporter. Nat Struct Mol Biol 22, 238–241 (2015). https://doi.org/10.1038/nsmb.2975
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2975
This article is cited by
-
Roles of adenine methylation in the physiology of Lacticaseibacillus paracasei
Nature Communications (2023)
-
Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter
Nature Communications (2023)
-
Transporters of glucose and other carbohydrates in bacteria
Pflügers Archiv - European Journal of Physiology (2020)
-
Structure of the mannose transporter of the bacterial phosphotransferase system
Cell Research (2019)
-
Insights into the molecular mechanism of dehalogenation catalyzed by D-2-haloacid dehalogenase from crystal structures
Scientific Reports (2018)