Abstract
The γ-tubulin ring complex (γTuRC) is the primary microtubule nucleator in cells. γγTuRC is assembled from repeating γγ-tubulin small complex (γTuSC) subunits and is thought to function as a template by presenting a γ-tubulin ring that mimics microtubule geometry. However, a previous yeast γTuRC structure showed γTuSC in an open conformation that prevents matching to microtubule symmetry. By contrast, we show here that γ-tubulin complexes are in a closed conformation when attached to microtubules. To confirm the functional importance of the closed γTuSC ring, we trapped the closed state and determined its structure, showing that the γ-tubulin ring precisely matches microtubule symmetry and providing detailed insight into γTuRC architecture. Importantly, the closed state is a stronger nucleator, thus suggesting that this conformational switch may allosterically control γTuRC activity. Finally, we demonstrate that γTuRCs have a strong preference for tubulin from the same species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Choy, R.M., Kollman, J.M., Zelter, A., Davis, T.N. & Agard, D.A. Localization and orientation of the γ-tubulin small complex components using protein tags as labels for single particle EM. J. Struct. Biol. 168, 571–574 (2009).
Kollman, J.M. et al. The structure of the γ-tubulin small complex: implications of its architecture and flexibility for microtubule nucleation. Mol. Biol. Cell 19, 207–215 (2008).
Oegema, K. et al. Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144, 721–733 (1999).
Zheng, Y., Wong, M.L., Alberts, B. & Mitchison, T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature 378, 578–583 (1995).
Kollman, J.M., Merdes, A., Mourey, L. & Agard, D.A. Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12, 709–721 (2011).
Moritz, M., Braunfeld, M.B., Guenebaut, V., Heuser, J. & Agard, D.A. Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2, 365–370 (2000).
Oakley, B.R., Oakley, C.E., Yoon, Y. & Jung, M.K. γ-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell 61, 1289–1301 (1990).
Kilmartin, J.V. & Goh, P.Y. Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J. 15, 4592–4602 (1996).
Sundberg, H.A. & Davis, T.N. A mutational analysis identifies three functional regions of the spindle pole component Spc110p in Saccharomyces cerevisiae. Mol. Biol. Cell 8, 2575–2590 (1997).
Kollman, J.M., Polka, J.K., Zelter, A., Davis, T.N. & Agard, D.A. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466, 879–882 (2010).
Tilney, L.G. et al. Microtubules: evidence for 13 protofilaments. J. Cell Biol. 59, 267–275 (1973).
O'Toole, E.T., Winey, M. & McIntosh, J.R. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10, 2017–2031 (1999).
Erlemann, S. et al. An extended γ-tubulin ring functions as a stable platform in microtubule nucleation. J. Cell Biol. 197, 59–74 (2012).
Sui, H. & Downing, K.H. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure 18, 1022–1031 (2010).
Keating, T.J. & Borisy, G.G. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat. Cell Biol. 2, 352–357 (2000).
Wiese, C. & Zheng, Y. A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat. Cell Biol. 2, 358–364 (2000).
Aldaz, H., Rice, L.M., Stearns, T. & Agard, D.A. Insights into microtubule nucleation from the crystal structure of human γ-tubulin. Nature 435, 523–527 (2005).
Rice, L.M., Montabana, E.A. & Agard, D.A. The lattice as allosteric effector: structural studies of αβ- and γ-tubulin clarify the role of GTP in microtubule assembly. Proc. Natl. Acad. Sci. USA 105, 5378–5383 (2008).
Egelman, E.H. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J. Struct. Biol. 157, 83–94 (2007).
Johnson, V., Ayaz, P., Huddleston, P. & Rice, L.M. Design, overexpression, and purification of polymerization-blocked yeast αβ-tubulin mutants. Biochemistry 50, 8636–8644 (2011).
Vinh, D.B., Kern, J.W., Hancock, W.O., Howard, J. & Davis, T.N. Reconstitution and characterization of budding yeast γ-tubulin complex. Mol. Biol. Cell 13, 1144–1157 (2002).
Choi, Y.K., Liu, P., Sze, S.K., Dai, C. & Qi, R.Z. CDK5RAP2 stimulates microtubule nucleation by the γ-tubulin ring complex. J. Cell Biol. 191, 1089–1095 (2010).
Löwe, J., Li, H., Downing, K.H. & Nogales, E. Refined structure of αβ-tubulin at 3.5 A resolution. J. Mol. Biol. 313, 1045–1057 (2001).
Ravelli, R.B. et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 428, 198–202 (2004).
Holt, L.J. et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 (2009).
Albuquerque, C.P. et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics 7, 1389–1396 (2008).
Holinger, E.P. et al. Budding yeast centrosome duplication requires stabilization of Spc29 via Mps1-mediated phosphorylation. J. Biol. Chem. 284, 12949–12955 (2009).
Keck, J.M. et al. A cell cycle phosphoproteome of the yeast centrosome. Science 332, 1557–1561 (2011).
Lin, T.C. et al. Phosphorylation of the yeast γ-tubulin Tub4 regulates microtubule function. PLoS ONE 6, e19700 (2011).
Vogel, J. et al. Phosphorylation of γ-tubulin regulates microtubule organization in budding yeast. Dev. Cell 1, 621–631 (2001).
Nazarova, E. et al. Distinct roles for antiparallel microtubule pairing and overlap during early spindle assembly. Mol. Biol. Cell 24, 3238–3250 (2013).
Samejima, I., Miller, V.J., Groocock, L.M. & Sawin, K.E. Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the γ-tubulin complex in vivo. J. Cell Sci. 121, 3971–3980 (2008).
Goshima, G., Mayer, M., Zhang, N., Stuurman, N. & Vale, R.D. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181, 421–429 (2008).
Rout, M.P. & Kilmartin, J.V. Yeast spindle pole body components. Cold Spring Harb. Symp. Quant. Biol. 56, 687–692 (1991).
Donaldson, A.D. & Kilmartin, J.V. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPD) with an essential function during SPB duplication. J. Cell Biol. 132, 887–901 (1996).
Zheng, Q.S., Braunfeld, M.B., Sedat, J.W. & Agard, D.A. An improved strategy for automated electron microscopic tomography. J. Struct. Biol. 147, 91–101 (2004).
Scheres, S.H.W., Melero, R., Valle, M. & Carazo, J.-M. Averaging of electron subtomograms and random conical tilt reconstructions through likelihood optimization. Structure 17, 1563–1572 (2009).
Förster, F., Pruggnaller, S., Seybert, A. & Frangakis, A.S. Classification of cryo-electron sub-tomograms using constrained correlation. J. Struct. Biol. 161, 276–286 (2008).
Sikorski, R.S. & Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 (1989).
Ohi, M., Li, Y., Cheng, Y. & Walz, T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 (2004).
Quispe, J. et al. An improved holey carbon film for cryo-electron microscopy. Microsc. Microanal. 13, 365–371 (2007).
Mindell, J.A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Egelman, E.H. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000).
Sachse, C. et al. High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. J. Mol. Biol. 371, 812–835 (2007).
Frank, J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies (Academic Press, San Diego, 1996).
Pettersen, E.F. et al. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Muller, E.G. A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol. Biol. Cell 7, 1805–1813 (1996).
Muller, E.G. et al. The organization of the core proteins of the yeast spindle pole body. Mol. Biol. Cell 16, 3341–3352 (2005).
Shimogawa, M.M., Widlund, P.O., Riffle, M., Ess, M. & Davis, T.N. Bir1 is required for the tension checkpoint. Mol. Biol. Cell 20, 915–923 (2009).
Pei, J., Kim, B.H. & Grishin, N.V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008).
Sali, A. & Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Trabuco, L.G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008).
Acknowledgements
The authors are grateful to M. Braunfeld and X. Li for assistance with cryo-EM data collection and L. Rice for help with overexpression and purification of yeast tubulin. This work was funded by the Howard Hughes Medical Institute and by US National Institutes of Health grants R01 GM031627 to D.A.A., R01 GM040506 to T.N.D. and T32 GM007270 to K.K.F.
Author information
Authors and Affiliations
Contributions
J.M.K. prepared samples, collected cryo-EM data, performed three-dimensional reconstructions, analyzed data and wrote the paper. C.H.G. performed molecular modeling, analyzed results and contributed to writing the paper. S.L. prepared samples, collected tomographic data, performed tomographic reconstructions and analyzed data. M.M. performed microtubule nucleation assays and analyzed data. A.Z. generated expression constructs and optimized protein expression. K.K.F. performed yeast viability assays and spindle morphology experiments and analyzed data. J.-J.F. assisted with subvolume averaging. A.S. analyzed data and contributed to writing the paper. J.K. provided samples and analyzed data. T.N.D. analyzed data and contributed to writing the paper. D.A.A. analyzed data and contributed to writing the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 The structure of wild-type γTuSC oligomers does not perfectly match 13-protofilament-microtubule geometry.
a) Two laterally-associated γTuSC subunits from ring-like γTuSC assemblies are shown. The two γ-tubulins (gold) within each γTuSC are held separate from each other by GCP2 and GCP3, while the γ-tubulins at the inter-γTuSC interface are positioned with microtubule-like lateral contacts. b) The intra-γTuSC separation results in a staggered pattern of γ-tubulin in γTuSC assemblies, with a helical pitch of 147 Å. c) By contrast, lateral contacts between tubulin subunits in a microtubule are uniform around the filament, and have a helical pitch of 122 Å. d) To illustrate the mismatch between γTuSC oligomers and the microtubule, the upper γ-tubulin in the ring has been aligned to make longitudinal contacts with α-tubulin at the minus end of the microtubule. This would result in a large gap between α- and γ-tubulin at the last position in the ring.
Supplementary Figure 2 γTuSCCC does not affect cell viability.
a) Representative images of WT, glr1Δ, and γTuSCCC glr1Δ fluorescent strains. GLR1 encodes glutathione reductase; glr1Δ cells were used to maximize the chance that the γTuSCCC disulfides would be oxidized. Cells in the top row have the spindle pole bodies marked by Spc42-mCherry and the microtubules marked by GFP-Tub1. Cells in the bottom row have the spindle pole bodies marked by Spc42-mCherry and the kinetochores marked by Nuf2-GFP. b) The doubling times of WT and γTuSCCC strains. c) Average GFP-Tub1 fluorescence distributions show that γTuSCCC has no effect on tubulin distribution across three spindle length classifications. d) Average Nuf2-GFP fluorescence distributions show that γTuSCCC has no effect on kinetochore clustering in metaphase spindles (1.28-1.59 µm), while shorter spindles show kinetochores cluster slightly closer to the spindle pole bodies. Error bars represent the standard error of the mean.
Supplementary Figure 3 Cross-linking and structural analysis of γTuSCCC.
a) γTuSCCC in the presence of 5 mM DTT is predominantly monomeric complex, although γTuSC pairs were frequently observed. b) Under non-reducing conditions γTuSCs spontaneously assemble into oligomers, even in the presence of 500 mM KCl, which prevents association of wildtype γTuSC. c) SDS-PAGE of γTuSCCC under reducing conditions (5 mM DTT), and non-reducing conditions (0 mM DTT). In the non-reducing condition γ-tubulin forms disulfide crosslinks, resulting in a ladder of 1-5 γ-tubulin chains. Formation of γ-tubulin oligomers greater than two chains indicates that crosslinks were formed at both inter-γTuSC and intra-γTuSC interfaces. d) Co-purified γTuSCCC and Spc1101–220 was dialyzed for 72 h. against an oxidizing buffer containing 1mM oxidized glutathione. On SDS-PAGE the reduced sample has the expected bands for Spc1101–220, γ-tubulin, and GCP2/GCP3, while in the unreduced sample extensive γ-tubulin crosslinking prevents its migration into the gel. This highly-cross-linked sample was used for cryo-EM imaging. e) Cryo-EM image of cross-linked γTuSCCC-Spc1101–220 filaments (same sample as in d). f) Segments of γTuSCCC filaments were compared to projections of the γTuSC filament in the open state and a preliminary reconstruction of the closed state. The histogram plots the fractional difference in correlation coefficient, with negative values representing a better match to the open state. Segments matching better to the open state were omitted from the final refinement of the closed filament structure. g) Fourier shell correlation (FSC) of the final γTuSCCC-Spc1101–220 reconstruction indicates a resolution of 6.9 Å at the 0.5 cutoff. Scalebars, 100 nm.
Supplementary Figure 4 Yeast γTuSC nucleation activity is species specific.
a) Comparison of nucleating activity of γTuSC rings with yeast and pig brain tubulin. Nucleation assays were performed as in Fig. 3C (data for yeast tubulin is the same as Fig. 3C), except that the experiments with brain tubulin were at 12 μM tubulin and 37° C (n=4 independent experiments; error bars represent the s.e.m.). b) The predicted longitudinal interaction between α- and γ-tubulin is shown at left. Human γ-tubulin and pig α-tubulin are shown as these are the existing crystal structures. At right the interaction surfaces are highlighted as a function of the conservation between yeast and vertebrate tubulin: grey sites are identical, yellow have conservative amino acid substitutions, red have non-conservative substitutions, magenta is a two amino acid insertion in γ-tubulin, and GTP is shown in orange. c) Binary sequence alignment of yeast and vertebrate a-tubulin. d) Binary sequence alignment of yeast and vertebrate g-tubulin. Color in g (c) and (d) is the same as for (b).
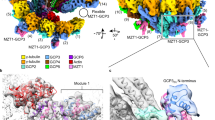
Supplementary Figure 5 Subunit localization and conformation in the γTuSC closed-state structure.
Comparison of the open and closed states of γTuSC. The elongated GCP2 and GCP3 structures each appear slightly straightened. b) Overlay of GCP3 in the open (grey) and closed (blue) states shows that it straightens by about 9°. c) Overlay of GCP2 in the open (grey) and closed (blue) states shows that it straightens by about 8°. d) Superposition of GCP2 and GCP3 in the closed state shows that they are in very similar overall conformation. GCP4, which was used as the starting structure for homology modeling both GCP2 and GCP3, is in a similar overall conformation. e) Superposition of GCP2 and GCP3 in the open state. For B-E all structural alignments were done using the two N-terminal helical bundles. f) The density predicted to correspond to part of Spc1101–220 resembles a two-stranded coiled-coil. A generic two-stranded coiled coil was used to model Spc110, and flexibly fit into the EM density. The closest contacts with Spc110 are between residues 77-97 of GCP2 (which includes 9 residues not included in our model) near the very base of γTuSC, and further up the structure with residues 119-126 of GCP3. g) A cutaway view of the γTuSCCC filament in the closed state, low-pass filtered at 15 Å. The γTuSC and modeled Spc110 structure is colored grey, and disordered density that runs along the center of the filament is colored green. Arrows denote the connections between the ends of the modeled Spc110 structure and the disordered density, which suggest that the disordered density is composed of flexible regions of Spc1101–220.
Supplementary Figure 6 Mapping phosphorylation sites on the γTuSC structure.
a) A single γTuSC is shown with residues known to be phosphorylated highlighted as spheres in red, yellow, and orange for sites on γ-tubulin, GCP2, and GCP3 respectively. c) The model of yeast γTuRC with known phosphorylation sites rendered as spheres. Sites freely accessible on the outer surface of the ring are colored purple, sites at the plus end of γ-tubulin predicted to be involved in longitudinal contacts with the microtubule in blue, and sites on the ring interior that would be inaccessible when a microtubule is bound are coral.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 10072 kb)
Capped microtubule minus end
The subtomogram average of capped microtubule ends from spindle pole bodies (MOV 5195 kb)
Comparison of γTuSC ring with capped microtubule end
A single ring of γTuSCCC-Spc1101–220 in the closed state is fit to the subtomogram average of capped microtubule ends from spindle pole bodies (MOV 6890 kb)
Model of γTuRC-microtubule interactions
A single ring of γTuSCCC-Spc1101–220 in the closed state, colored by subunit, is shown followed by the model of longitudinal interactions with a microtubule. (MOV 14930 kb)
Rights and permissions
About this article
Cite this article
Kollman, J., Greenberg, C., Li, S. et al. Ring closure activates yeast γTuRC for species-specific microtubule nucleation. Nat Struct Mol Biol 22, 132–137 (2015). https://doi.org/10.1038/nsmb.2953
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2953
This article is cited by
-
Mechanisms underlying spindle assembly and robustness
Nature Reviews Molecular Cell Biology (2023)
-
Centrosome: A Microtubule Nucleating Cellular Machinery
Journal of the Indian Institute of Science (2021)
-
Directing curli polymerization with DNA origami nucleators
Nature Communications (2019)
-
Autonomous helical propagation of active toroids with mechanical action
Nature Communications (2019)
-
Separation and Loss of Centrioles From Primordidal Germ Cells To Mature Oocytes In The Mouse
Scientific Reports (2018)