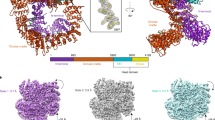
Abstract
Mammalian CtIP protein has major roles in DNA double-strand break (DSB) repair. Although it is well established that CtIP promotes DNA-end resection in preparation for homology-dependent DSB repair, the molecular basis for this function has remained unknown. Here we show by biophysical and X-ray crystallographic analyses that the N-terminal domain of human CtIP exists as a stable homotetramer. Tetramerization results from interlocking interactions between the N-terminal extensions of CtIP's coiled-coil region, which lead to a 'dimer-of-dimers' architecture. Through interrogation of the CtIP structure, we identify a point mutation that abolishes tetramerization of the N-terminal domain while preserving dimerization in vitro. Notably, we establish that this mutation abrogates CtIP oligomer assembly in cells, thus leading to strong defects in DNA-end resection and gene conversion. These findings indicate that the CtIP tetramer architecture described here is essential for effective DSB repair by homologous recombination.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Jackson, S.P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Krejci, L., Altmannova, V., Spirek, M. & Zhao, X. Homologous recombination and its regulation. Nucleic Acids Res. 40, 5795–5818 (2012).
Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010).
Fusco, C., Reymond, A. & Zervos, A.S. Molecular cloning and characterization of a novel retinoblastoma-binding protein. Genomics 51, 351–358 (1998).
Schaeper, U., Subramanian, T., Lim, L., Boyd, J.M. & Chinnadurai, G. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J. Biol. Chem. 273, 8549–8552 (1998).
Wong, A.K. et al. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene 17, 2279–2285 (1998).
Li, S. et al. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature 406, 210–215 (2000).
Chen, P.L. et al. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 25, 3535–3542 (2005).
Liu, F. & Lee, W.-H. CtIP activates its own and cyclin D1 promoters via the E2F/RB pathway during G1/S progression. Mol. Cell. Biol. 26, 3124–3134 (2006).
Yu, X. & Chen, J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 24, 9478–9486 (2004).
Sartori, A.A. et al. Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007).
Yun, M.H. & Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459, 460–463 (2009).
Huertas, P. et al. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692 (2008).
Zhang, Y. & Jasin, M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat. Struct. Mol. Biol. 18, 80–84 (2011).
Lee-Theilen, M., Matthews, A.J., Kelly, D., Zheng, S. & Chaudhuri, J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat. Struct. Mol. Biol. 18, 75–79 (2011).
You, Z. et al. CtIP links DNA double-strand break sensing to resection. Mol. Cell 36, 954–969 (2009).
Huertas, P. & Jackson, S.P. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565 (2009).
Yuan, J. & Chen, J. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J. Biol. Chem. 284, 31746–31752 (2009).
Dubin, M.J. et al. Dimerization of CtIP, a BRCA1- and CtBP-interacting protein, is mediated by an N-terminal coiled-coil motif. J. Biol. Chem. 279, 26932–26938 (2004).
Stokes, P.H., Thompson, L.S., Marianayagam, N.J. & Matthews, J.M. Dimerization of CtIP may stabilize in vivo interactions with the Retinoblastoma-pocket domain. Biochem. Biophys. Res. Commun. 354, 197–202 (2007).
Wang, H. et al. CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J. Biol. Chem. 287, 21471–21480 (2012).
Säbel, C.E., Shepherd, J.L. & Siemann, S. A direct spectrophotometric method for the simultaneous determination of zinc and cobalt in metalloproteins using 4-(2-pyridylazo)resorcinol. Anal. Biochem. 391, 74–76 (2009).
Certo, M.T. et al. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods 8, 671–676 (2011).
Gomez-Cabello, D., Jimeno, S., Fernández-Ávila, M.J. & Huertas, P. New tools to study DNA double-strand break repair pathway choice. PLoS ONE 8, e77206 (2013).
McVey, M. & Lee, S.E. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 24, 529–538 (2008).
Sharma, A., Singh, K. & Almasan, A. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol. Biol. 920, 613–626 (2012).
Huertas, P. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 17, 11–16 (2010).
Forment, J.V., Walker, R.V. & Jackson, S.P. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytometry A 81, 922–928 (2012).
Pommier, Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 (2006).
Saleh-Gohari, N. et al. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 25, 7158–7169 (2005).
Makharashvili, N. et al. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol. Cell 54, 1022–1033 (2014).
Wang, H. et al. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell 54, 1012–1021 (2014).
Hopfner, K.-P. et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418, 562–566 (2002).
Lobachev, K.S., Gordenin, D.A. & Resnick, M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108, 183–193 (2002).
Clerici, M., Mantiero, D., Lucchini, G. & Longhese, M.P. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 280, 38631–38638 (2005).
Lengsfeld, B.M., Rattray, A.J., Bhaskara, V., Ghirlando, R. & Paull, T.T. Sae2 Is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28, 638–651 (2007).
Fu, Q. et al. Phosphorylation-regulated transitions in an oligomeric state control the activity of the Sae2 DNA repair enzyme. Mol. Cell. Biol. 34, 778–793 (2014).
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 (2004).
Peränen, J., Rikkonen, M., Hyvonen, M. & Kaariainen, L. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236, 371–373 (1996).
Sreerama, N. & Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 (2000).
Whitmore, L. & Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89, 392–400 (2008).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Collaborative Computational Project, N. 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Kelley, L.A. & Sternberg, M.J.E. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 (2009).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Dignam, J.D., Lebovitz, R.M. & Roeder, R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 (1983).
Richardson, C., Moynahan, M.E. & Jasin, M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12, 3831–3842 (1998).
Limoli, C.L. & Ward, J.F. A new method for introducing double-strand breaks into cellular DNA. Radiat. Res. 134, 160–169 (1993).
Bekker-Jensen, S. et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173, 195–206 (2006).
Acknowledgements
We thank M. Kilkenny for help with the collection of X-ray diffraction data, A. Sharff and P. Keller for help with X-ray data processing and J.D. Maman for assistance with SEC-MALS. This work was supported by a Wellcome Trust Senior Research Fellowship award in basic biomedical sciences (L.P.), an Isaac Newton Trust research grant (L.P. and O.R.D.) and a Cambridge Overseas Trust PhD studentship (M.D.S.). Research in the laboratory of S.P.J. is funded by Cancer Research UK (CRUK; programme grant C6/A11224), the European Research Council and the European Community Seventh Framework Programme (grant agreement no. HEALTH-F2-2010-259893 (DDResponse)). Core funding is provided by Cancer Research UK (C6946/A14492) and the Wellcome Trust (WT092096). S.P.J. receives his salary from the University of Cambridge, supplemented by CRUK. J.V.F. is funded by Cancer Research UK programme grant C6/A11224 and the Ataxia Telangiectasia Society. R.B. and J.C. are funded by Cancer Research UK programme grant C6/A11224. Y.G. and M.D. are funded by the European Research Council grant DDREAM.
Author information
Authors and Affiliations
Contributions
O.R.D. and M.S. expressed and purified the CtIP proteins and performed the biochemical and biophysical experiments; C.R.M. and N.J.R. performed the electrophoretic mobility shift assays; O.R.D. determined the X-ray crystal structure of CtIP-nNTD; J.V.F. performed the TLR and MMEJ assays, the DNA-end resection assays, the immunofluorescence assays (with help from Y.G.), the GFP-CtIP quantification on fluorescence-activated cell sorting and the coimmunoprecipitation experiments; R.B. performed the gel-filtration analysis of Flag-CtIP from HEK-293T cell extracts and created the Flag-tagged CtIP constructs; J.C. established the U2OS-TLR and the U2OS-MMEJ stable cell lines; M.D. established the U2OS GFP-CtIP cell lines; J.V.F. and Y.G. established the TLR system in U2OS cells; O.R.D., J.V.F., S.P.J. and L.P. designed experiments and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Multiple sequence alignment of the CtIP N-terminal domain, purified CtIP protein constructs and details of the 2Fo – Fc electron density map of CtIP-NTD.
(a) Multiple sequence alignment, including the predicted secondary structure (red rods indicate regions of predicted α-helix) and coiled-coil regions; the probability of coiled-coil formation is indicated by ‘-’ (<50%), ‘c’ (50-90%) or ‘C’ (>90%). Protein sequence analysis was performed with the Jalview suite of bioinformatics tools (www.jalview.org). Hs: Homo sapiens, Mm: Mus musculus, Ec: Equus caballus, Oc: Oryctolagus cuniculus, Gg: Gallus gallus, Xl: Xenopus laevis, Dr: Danio rerio. (b) SDS-PAGE analysis of purified recombinant CtIP protein samples: NTD, NTD-DM (C89A, C92A), nNTD, cNTD, cNTD-DM (C89A, C92A) and CTD visualised by Coomassie staining. (c) Details of the 2Fo - Fc electron density map, contoured at 1.2 rmsd, superimposed on the refined crystallographic model of CtIP-NTD. The figure shows the map in the vicinity of amino acid W24. The map is shown as thin mesh in light purple, the protein atoms are shown in ball-and-stick representation and coloured according to chemical identity. The figure was prepared in Coot
Supplementary Figure 2 CD analysis of CtIP N-terminal-domain constructs and microPIXE analysis of the CtIP N-terminal domain.
Circular dichroism (CD) analysis of CtIP N-terminal domain constructs. (a, c, e) Far-UV CD spectra recorded between 260 and 185 nm, in mean residue ellipticity, MRE ([θ]) (1000x deg.cm2.dmol-1.residue-1), with α-helical content calculated through deconvolution using the CDSSTR algorithm with normalised root-mean-square deviation values (nrmsd) as shown. (b, d, f) CD thermal denaturation data recorded between 5 and 95°C, in mean residue ellipticity at 222 nm ([θ]222). (a) CD spectra and (b) thermal denaturation of CtIP-NTD (solid line) and CtIP-NTD-DM (dashed line). (c) CD spectra and (d) thermal denaturation of CtIP-cNTD (solid line) and CtIP-cNTD-DM (dashed line). (e) CD spectra and (f) thermal denaturation of CtIP-nNTD. (g) CtIP-NTD was analysed in buffer containing KBr. Rutherford backscattering (RBS) spectrum and (h) PIXE spectrum with raw data shown as black dots and the modelling fitting as red lines. (i) Zinc was detected at a mean atomic ratio to sulphur of 0.095, which given the presence of three methionine and three cysteine residues per chain, corresponds to a CtIP-NTD:Zn2+ ratio of 1.8:1.
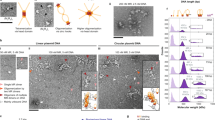
Supplementary Figure 3 SEC-MALS analysis of MBP-CtIP-nNTD and F20E CtIP-NTD.
(a) MBP-CtIP-nNTD eluted in a single peak corresponding to a molecular weight of 187 kDa; its theoretical tetramer size is 197 kDa. For comparison, free MBP eluted in a single peak of 44.5 kDa, corresponding to its theoretical monomer size of 44.7 kDa. The light scattering (LS) as relative Raleigh ratio and the differential Reflective Index (dRI) are drawn as solid and dashed lines, respectively. The value for the fitted molecular mass (Mr) of MBP-CtIP-nNTD is shown as diamond shapes across its elution peak. The predicted molecular masses of monomeric MBP-CtIP-nNTD and free MBP are shown in brackets above the respective elution peak. (b) F20E CtIP-NTD forms two distinct species of 62.3 kDa and 33.2 kDa, corresponding to its theoretical tetramer and dimer sizes of 63.2 kDa and 31.6 kDa, respectively. The light scattering (LS) as relative Raleigh ratio and the differential Reflective Index (dRI) are drawn as solid and dashed lines, respectively. The values for the fitted molecular masses (Mr) of dimeric and tetrameric F20E CtIP-NTD are shown as diamond shapes across the respective elution peaks. The predicted molecular mass of monomeric F20E CtIP-nNTD is shown in brackets above the elution peaks.
Supplementary Figure 4 Experimental procedure and gating scheme for the TLR system.
(a) TLR cells are not fluorescent due to the fact that the TLR cassette contains an in-frame truncated version of the eGFP gene and full-length mCherry is out of frame. When TLR cells are transfected with an I-SceI nuclease encoded in a plasmid also expressing an infrared fluorescent protein (IFP) and an exogenous donor template containing the missing part of eGFP that also expresses a blue fluorescent protein (BFP), the different repair outcomes will give different fluorescent cells. If the double-strand break (DSB) is repaired by gene conversion (homologous recombination), eGFP is restored and the cells fluoresce in green. If the DSB is repaired by mutagenic end joining causing a +2 frameshift, mCherry gets in frame and its expression yields red fluorescence. The T2A “dis-linker” between eGFP and mCherry allows the downstream-encoded mCherry to escape degradation of the misfolded protein encoded in the +3 reading frame of eGFP. (b) Experimental outline of the TLR assay. (c-h) Gating scheme for a TLR assay. (c) Discrimination of cells over debris using a forward scatter vs. side scatter plot (areas). (d) Discrimination of cell singlets over doublets using a forward scatter (area) vs. side scatter (width) plot. (e) Selection of events for quantification. Dot plot showing the intensity of BFP (donor; x axis) vs. intensity of IFP (nuclease; y axis). Cells with negative staining for both BFP and IFP are depicted in (f) to show the absence of eGFP or mCherry fluorescence in non-transfected cells, and to establish quantification gates. Cells counted positive for both BFP and IFP (that is, transfected with both the donor and the nuclease) are represented in (g), where quantification gates are the same as the ones established in (f). At least 10,000 events were analysed in the double-positive population. (h) Due to the fact that the amount of donor transfected in the cells affects the outcome of the assay, the intensity of the BFP signal in the double-positive cells (geometric mean of BFP signal in ‘BFP+ IFP+’ cells) was calculated for normalisation purposes (see Methods). (i) Mutagenic end-joining (mutEJ) rates in the TLR-CtIP complementation system. Only expression of the wild-type version of FLAG-CtIP reduced the rate of mutEJ to values similar to the control (cells transfected with control siRNA). EV: empty vector. All quantifications are shown as the average of three independent experiments. Error bars are ± SEM.
Supplementary Figure 5 CtIP expression levels in the different stable cell lines generated and fluorescence-activated cell sorting quantification of GFP-CtIP accumulation on damaged cells.
(a) CtIP expression levels in the different U2OS-MMEJ FLAG-CtIP clones. Samples were collected 48 h after siRNA transfection. CtIP antibody was a gift from R. Baer. PARP-1 antibody (Cell Signalling) was used as loading control. (b) GFP-CtIP expression levels in the U2OS stable cell lines. RPA34-20 (Merck) antibody was used as loading control. (c) A representative complete dataset for quantification of GFP-CtIP accumulation on damaged cells. Dot plots representing, for each dataset, gating and quantification (in the y axis) of the GFP-positive cells (top panels) and the γH2AX-positive cells (bottom panels) taking into account their DNA content (x axis). Quantification gates were always established using the untreated samples of each dataset. At least 10,000 events were analysed per sample.
Supplementary Figure 6 A representative complete dataset for the DNA-end resection assay.
Dot plots representing, for each dataset, gating and quantification (in the y axis) of the RPA-positive cells (top panels) and the γH2AX-positive cells (bottom panels) taking into account their DNA content (x axis). Quantification gates were always established using the untreated samples of each dataset and were defined as to avoid quantification of RPA chromatinization due to DNA replication. Note that as camptothecin (CPT) only causes DNA double-strand breaks in S-phase cells, both RPA- and γH2AX-positive cells only appear in that cell cycle stage. At least 10,000 events were analyzed per sample.
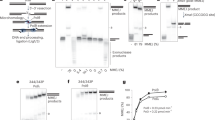
Supplementary Figure 7 CtIP-CTD, but not CtIP-NTD, interacts with double-stranded DNA.
Electrophoretic mobility shift assay measuring the ability of CtIP NTD and CTD regions to interact with linear, double-stranded DNA. In the experiment, increasing amounts (0, 0.5, 1.0, 2.0, 5.0 and 10μM) of the CtIP protein were incubated with a 1μM sample of 5’ fluorescein-labeled 200bp double-stranded DNA. After incubation, each sample was resolved by electrophoresis on agarose gel and visualised under UV light.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 5362 kb)
Supplementary Data Set 1
Uncropped versions of western blots presented in Figures 4 and 6. (PDF 7301 kb)
Rights and permissions
About this article
Cite this article
Davies, O., Forment, J., Sun, M. et al. CtIP tetramer assembly is required for DNA-end resection and repair. Nat Struct Mol Biol 22, 150–157 (2015). https://doi.org/10.1038/nsmb.2937
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2937
This article is cited by
-
Recurrence mutation in RBBP8 gene causing non-syndromic autosomal recessive primary microcephaly; geometric simulation approach for insight into predicted computational models
Journal of Human Genetics (2023)
-
Stereo- and regiodefined DNA-encoded chemical libraries enable efficient tumour-targeting applications
Nature Chemistry (2021)
-
The role of poly(ADP-ribose) polymerase inhibitors in the treatment of cancer and methods to overcome resistance: a review
Cell & Bioscience (2020)
-
Prognosis value of RBBP8 expression in plasma cell myeloma
Cancer Gene Therapy (2020)
-
Regulatory control of DNA end resection by Sae2 phosphorylation
Nature Communications (2018)