Abstract
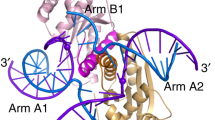
The enzymatic hydrolysis of DNA phosphodiester bonds has been widely studied, but the chemical reaction has not yet been observed. Here we follow the generation of a DNA double-strand break (DSB) by the Desulfurococcus mobilis homing endonuclease I-DmoI, trapping sequential stages of a two-metal-ion cleavage mechanism. We captured intermediates of the different catalytic steps, and this allowed us to watch the reaction by 'freezing' multiple states. We observed the successive entry of two metals involved in the reaction and the arrival of a third cation in a central position of the active site. This third metal ion has a crucial role, triggering the consecutive hydrolysis of the targeted phosphodiester bonds in the DNA strands and leaving its position once the DSB is generated. The multiple structures show the orchestrated conformational changes in the protein residues, nucleotides and metals during catalysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Roberts, R.J. How restriction enzymes became the workhorses of molecular biology. Proc. Natl. Acad. Sci. USA 102, 5905–5908 (2005).
Pingoud, V. et al. On the divalent metal ion dependence of DNA cleavage by restriction endonucleases of the EcoRI family. J. Mol. Biol. 393, 140–160 (2009).
Roberts, R.J. & Halford, S.E. in Nucleases (eds. Linn, S.M., Lloyd, R.S. & Roberts, R.J.) 35–88 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1993).
Gerlt, J.A., Coderre, J.A. & Mehdi, S. Oxygen chiral phosphate esters. Adv. Enzymol. 55, 291–380 (1983).
Steitz, T.A. & Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA 90, 6498–6502 (1993).
Yang, W., Lee, J.Y. & Nowotny, M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 (2006).
Yang, W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 44, 1–93 (2011).
Marcaida, M.J., Munoz, I.G., Blanco, F.J., Prieto, J. & Montoya, G. Homing endonucleases: from basics to therapeutic applications. Cell. Mol. Life Sci. 67, 727–748 (2010).
Stoddard, B.L. Homing endonuclease structure and function. Q. Rev. Biophys. 38, 49–95 (2005).
Arnould, S. et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 355, 443–458 (2006).
Ashworth, J. et al. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature 441, 656–659 (2006).
Chen, Z., Wen, F., Sun, N. & Zhao, H. Directed evolution of homing endonuclease I-SceI with altered sequence specificity. Protein Eng. Des. Sel. 22, 249–256 (2009).
Rosen, L.E. et al. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 34, 4791–4800 (2006).
Muñoz, I.G. et al. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 39, 729–743 (2011).
Redondo, P. et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature 456, 107–111 (2008).
Takeuchi, R. et al. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc. Natl. Acad. Sci. USA 108, 13077–13082 (2011).
Kjems, J. & Garrett, R.A. An intron in the 23S ribosomal gene of the archaebacterium Desulfurococcus mobilis. Nature 318, 675–677 (1985).
Dalgaard, J.Z., Garrett, R.A. & Belfort, M. A site-specific endonuclease encoded by a typical archaeal intron. Proc. Natl. Acad. Sci. USA 90, 5414–5417 (1993).
Dalgaard, J.Z., Garrett, R.A. & Belfort, M. Purification and characterization of two forms of I-DmoI, a thermophilic site-specific endonuclease encoded by an archaeal intron. J. Biol. Chem. 269, 28885–28892 (1994).
Chevalier, B.S. & Stoddard, B.L. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 29, 3757–3774 (2001).
Marcaida, M.J. et al. Crystal structure of I-DmoI in complex with its target DNA provides new insights into meganuclease engineering. Proc. Natl. Acad. Sci. USA 105, 16888–16893 (2008).
Bolduc, J.M. et al. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 17, 2875–2888 (2003).
Chevalier, B. et al. Metal-dependent DNA cleavage mechanism of the I-CreI LAGLIDADG homing endonuclease. Biochemistry 43, 14015–14026 (2004).
Moure, C.M., Gimble, F.S. & Quiocho, F.A. The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity. J. Mol. Biol. 334, 685–695 (2003).
Freudenthal, B.D., Beard, W.A., Shock, D.D. & Wilson, S.H. Observing a DNA polymerase choose right from wrong. Cell 154, 157–168 (2013).
Nakamura, T., Zhao, Y., Yamagata, Y., Hua, Y.J. & Yang, W. Watching DNA polymerase η make a phosphodiester bond. Nature 487, 196–201 (2012).
Prieto, J. et al. Generation and analysis of mesophilic variants of the thermostable archaeal I-DmoI homing endonuclease. J. Biol. Chem. 283, 4364–4374 (2008).
Pontius, B.W., Lott, W.B. & von Hippel, P.H. Observations on catalysis by hammerhead ribozymes are consistent with a two-divalent-metal-ion mechanism. Proc. Natl. Acad. Sci. USA 94, 2290–2294 (1997).
Liao, R.-Z., Yu, J.-G. & Himo, F. Phosphate mono- and diesterase activities of the trinuclear zinc enzyme nuclease P1: insights from quantum chemical calculations. Inorg. Chem. 49, 6883–6888 (2010).
Ivanov, I., Tainer, J. & McCammon, J. Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV. Proc. Natl. Acad. Sci. USA 104, 1465–1470 (2007).
Chevalier, B.S. et al. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell 10, 895–905 (2002).
Gelpí, J.L. et al. Classical molecular interaction potentials: improved setup procedure in molecular dynamics simulations of proteins. Proteins 45, 428–437 (2001).
Borrelli, W.K., Vitalis, A., Alcantara, R. & Guallar, V. PELE: protein energy landscape exploration: a novel Monte Carlo based technique. J. Chem. Theory Comput. 1, 1304–1311 (2005).
Moure, C.M., Gimble, F.S. & Quiocho, F.A. Crystal structures of I-SceI complexed to nicked DNA substrates: snapshots of intermediates along the DNA cleavage reaction pathway. Nucleic Acids Res. 36, 3287–3296 (2008).
Gómez, H., Polyak, I., Thiel, W., Lluch, J.M. & Masgrau, L. Retaining glycosyltransferase mechanism studied by QM/MM methods: lipopolysaccharyl-α-1,4-galactosyltransferase C transfers α-galactose via an oxocarbenium ion-like transition state. J. Am. Chem. Soc. 134, 4743–4752 (2012).
Gómez, H., Lluch, J.M. & Masgrau, L. Substrate-assisted and nucleophilically assisted catalysis in bovine α-1,3-galactosyltransferase: mechanistic implications for retaining glycosyltransferases. J. Am. Chem. Soc. 135, 7053–7063 (2013).
Gómez, H. et al. Computational and experimental study of O-glycosylation: catalysis by human UDP-GalNAc polypeptide:GalNAc transferase-T2. Org. Biomol. Chem. 12, 2645–2655 (2014).
Rosta, E., Nowotny, M., Yang, W. & Hummer, G. Catalytic mechanism of RNA backbone cleavage by ribonuclease H from quantum mechanics/molecular mechanics simulations. J. Am. Chem. Soc. 133, 8934–8941 (2011).
Makoto, K., Haruki, N. & Yu, T. Density functional study of the phosphate diester hydrolysis of RNA in RNA/DNA hybrid by RNase HI. Mol. Phys. 112, 355–364 (2014).
De Vivo, M., Dal Peraro, M. & Klein, M. Phosphodiester cleavage in ribonuclease H occurs via an associative two-metal-aided catalytic mechanism. J. Am. Chem. Soc. 130, 10955–10962 (2008).
Elsässer, B. & Fels, G. Atomistic details of the associative phosphodiester cleavage in human ribonuclease H. Phys. Chem. Chem. Phys. 12, 11081–11088 (2010).
Ding, X., Rasmussen, B.F., Petsko, G.A. & Ringe, D. Direct structural observation of an acyl-enzyme intermediate in the hydrolysis of an ester substrate by elastase. Biochemistry 33, 9285–9293 (1994).
Ding, X., Rasmussen, B.F., Petsko, G.A. & Ringe, D. Direct crystallographic observation of an acyl-enzyme intermediate in the elastase-catalyzed hydrolysis of a peptidyl ester substrate: exploiting the “glass transition” in protein dynamics. Bioorg. Chem. 34, 410–423 (2006).
Murray, J.B., Szoke, H., Szoke, A. & Scott, W.G. Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction. Mol. Cell 5, 279–287 (2000).
Scott, W.G., Murray, J.B., Arnold, J.R., Stoddard, B.L. & Klug, A. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science 274, 2065–2069 (1996).
Redondo, P., Prieto, J., Ramos, E., Blanco, F.J. & Montoya, G. Crystallization and preliminary X-ray diffraction analysis on the homing endonuclease I-Dmo-I in complex with its target DNA. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 1017–1020 (2007).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Halford, S.E., Johnson, N.P. & Grinsted, J. The EcoRI restriction endonuclease with bacteriophage λ DNA: kinetic studies. Biochem. J. 191, 581–592 (1980).
Gordon, J.C. et al. H.: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33, W368–W371 (2005).
Myers, J., Grothaus, G., Narayanan, S. & Onufriev, A. A simple clustering algorithm can be accurate enough for use in calculations of pKs in macromolecules. Proteins 63, 928–938 (2006).
Anandakrishnan, R., Aguilar, B. & Onufriev, A. H. 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 40, W537–W541 (2012).
Sherwood, P. et al. QUASI: A general purpose implementation of the QM/MM approach and its application to problems in catalysis. J. Mol. Struct. Theochem. 632, 1–28 (2003).
Ahlrichs, R., Bär, M., Häser, M., Horn, H. & Kölmel, C. Electronic structure calculations on workstation computers: the program system turbomole. Chem. Phys. Lett. 162, 165–169 (1989).
Slater, J.C. A simplification of the Hartree-Fock method. Phys. Rev. 81, 385–390 (1951).
Becke, A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A 38, 3098–3100 (1988).
Becke, A.D. A new mixing of Hartree-Fock and local density-functional theories. J. Chem. Phys. 98, 1372–1377 (1993).
Stephens, P.J., Devlin, F.J., Chabalowski, C.F. & Frisch, M.J. Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 98, 11623–11627 (1994).
Lee, C., Yang, W. & Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter 37, 785–789 (1988).
Schäfer, A., Horn, H. & Ahlrichs, R. Fully optimized contracted Gaussian basis sets for atoms Li to Kr. J. Chem. Phys. 97, 2571–2577 (1992).
Clark, T., Chandrasekhar, J., Spitznagel, G.W. & Schleyer, P.V.R. Efficient diffuse function-augmented basis sets for anion calculations. III. The 3–21+G basis set for first-row elements, Li–F. J. Comput. Chem. 4, 294–301 (1983).
Smith, W. & Forester, T. DL_POLY_2.0: a general-purpose parallel molecular dynamics simulation package. J. Mol. Graph. 14, 136–141 (1996).
Scott, J.W. et al. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 106, 765–784 (1984).
Dirk, B. & Walter, T. Hybrid models for combined quantum mechanical and molecular mechanical approaches. J. Phys. Chem. 100, 10580–10594 (1996).
Sherwood, P. et al. Computer simulation of zeolite structure and reactivity using embedded cluster methods. Faraday Discuss. 106, 79–92 (1997).
Nocedal, J. Updating quasi-Newton matrices with limited storage. Math. Comput. 35, 773–782 (1980).
Banerjee, A., Adams, N., Simons, J. & Shepard, R. Search for stationary points on surfaces. J. Phys. Chem. 89, 52–57 (1985).
Baker, J. An algorithm for the location of transition states. J. Comput. Chem. 7, 385–395 (1986).
Billeter, S.R., Turner, A.J. & Thiel, W. Linear scaling geometry optimisation and transition state search in hybrid delocalised internal coordinates. Phys. Chem. Chem. Phys. 2, 2177–2186 (2000).
Baker, N.A., Sept, D., Joseph, S., Holst, M.J. & McCammon, J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98, 10037–10041 (2001).
Hernández-Ortega, A. et al. Substrate diffusion and oxidation in GMC oxidoreductases: an experimental and computational study on fungal aryl-alcohol oxidase. Biochem. J. 436, 341–350 (2011).
Hosseini, A. et al. Atomic picture of ligand migration in toluene 4-monooxygenase. J. Phys. Chem. B (10.1021/jp502509a) (13 May 2014).
Lucas, M.F. & Guallar, V. An atomistic view on human hemoglobin carbon monoxide migration processes. Biophys. J. 102, 887–896 (2012).
Lucas, M.F. & Guallar, V. Single vs. multiple ligand pathways in globins: a computational view. Biochim. Biophys. Acta 1834, 1739–1743 (2013).
Spyrakis, F. et al. Comparative analysis of inner cavities and ligand migration in non-symbiotic AHb1 and AHb2. Biochim. Biophys. Acta 1834, 1957–1967 (2013).
Hornak, V. et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 (2006).
Pérez, A. et al. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 92, 3817–3829 (2007).
Onufriev, A., Bashford, D. & Case, D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 55, 383–394 (2004).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Acknowledgements
We thank the Swiss Light Source (SLS) and ALBA beamline staff for their support. This work was supported by the Ministerio de Economía y Competitividad of Spain (BFU2011-23815/BMC to G.M.), the Novo Nordisk Foundation (grant NNF14CC0001 to G.M.), the Fundación Ramón Areces (to G.M.), the Comunidad Autónoma de Madrid (CAM-S2010/BMD-2305 to G.M.), the European Union Marie Curie 'SMARTBREAKER' (2010-276953 to S.S.), the Ministerio de Educación of Spain (SB2010-0105 to S.S.) and the Ministerio de Economia y Competitividad of Spain (JCI-2011-09308 to R.M. and BIO2012-32868 and ERC-SimDNA to M.O.).
Author information
Authors and Affiliations
Contributions
R.M. and S.S. performed the catalytic time-course experiment. P.R. prepared the crystals and performed the biochemical experiments. R.M. performed the crystallographic analysis and refined the data. M.J.M. refined some of the data sets. H.G. analyzed the data and performed the theoretical calculations. M.O. supervised the theoretical calculations and helped to write the manuscript. J.P. discussed the data, performed biochemical experiments and helped to write the manuscript. G.M. designed the crystallographic time-course experiment, helped with data processing and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
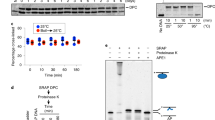
Supplementary Figure 1 Kinetics of I-DmoI cleavage under single-turnover conditions.
A) pH 8/65ºC and B) pH6/40ºC. First-order rate constants (k*) are plotted against I-DmoI initial concentrations ([I-DmoI]0). The correlation coefficients (R) corresponding to the curve fitting to the equation describing k* as a function of [I-DmoI]0 and the estimated kinetic parameters k*max and K M* are shown in the chart. One gel for each assay is presented as an example of the experimental data.
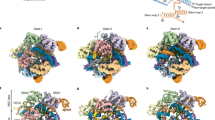
Supplementary Figure 2 Superposition of the active centers of I-DmoI in the presence of Mg2+ or Mn2+.
Each structure is colored according to the nature of the present divalent ions. A) The initial state of the reaction previously obtained in the presence of the non-catalytic metal Ca2+ (PDB Code 2VS7) is superimposed with the initial experimental state obtained in this manuscript. The rmsd (root mean square deviation) of the atoms in the active centre is 0.2 Å. B) Detailed view of the active centre comparing the conformation of the state 2. The DNA phosphodiester bonds are still intact and sites A and B are occupied with the metal, Mg2+ or Mn2+ depending on the soaking. The rmsd of the atoms in the active centre is 0.1 Å. C) A similar detailed view of the enzyme catalytic centre in state 7. This is the final state where both DNA strands are cleaved in the same way independently of the metal used in the experiment (Mg2+ or Mn2+), and the metal is found in sites A and B. The rmsd of the atoms in the active centre is 0.1 Å. The Mg2+ structures were solved at 2.20 and 2.35 Å resolution for states 2 and 7 respectively.
Supplementary Figure 3 PELE simulation results: interaction energies (kcal/mol) versus distance (Å) for the Mn2+ ions’ entrance along the channel formed in the I-DmoI–DNA interface.
A different colour is used for the interaction energies obtained for the independent 127 PELE trajectories. Distances are relative to reference positions (i.e. in PDB structures). Positions of binding sites A, B and C are highlighted with blue circles. Graphs correspond to: A-B) modelling of Mn2+ binding to site A when no ions are previously located in the active site; C-D), modelling of Mn2+ binding to site B when no ions are previously bound in the active site; E-F), modelling of Mn2+ binding to site B when there is an ion bound in site A and G-H), modelling of Mn2+ binding to site C when there are ions bound in sites A and B. The green and grey spheres in Figures B, D, F. H are used to show the reference positions and the places visited by the Mn2+ ions throughout the PELE ‘effective’ trajectories (.i.e. corresponding to simulations where the ions accessed the active site).
Supplementary Figure 4 PB calculations in the structure corresponding to the first stage of the catalytic process of I-DmoI.
A) Electrostatic potential visualization of the surface in complex I-DmoI/DNA. The arrow indicates the aperture of the channel in the protein/DNA interface where site A is located. The DNA double helix is highlighted in yellow. B) Calculated Molecular Interaction Potential (MIP) using Mn2+ as probe for the structure corresponding to the initial stage of catalysis (state 1). The black mesh corresponds to the calculation assuming that no divalent ions are bound to I-DmoI. The red mesh corresponds to the calculation assuming that Mn2+ ions were previously bound to sites A and B. The 100 points of highest interaction energy (electrostatic + Van der Waals) are depicted in each case. The Mn2+ atoms are represented as black spheres and the residues involved in their coordination are shown. Notice that site C is predicted to bind Mn2+ even when divalent ions are previously bound to sites A and B. Panels C and D depict the Potential Energy Surface (PES) at the QM(BP86/SVP)/MM(AMBER) level of theory corresponding to the movement of a divalent ion from site A or B to C. C) The divalent ion was modelled as Mg2+. The 90 atoms considered in the QM partition are highlighted. The starting structure in the calculations is depicted in D), where black and red arrows indicate the displacements modelled from site A to C or B to C, respectively. Note that only one ion was present in the calculations, which is initially bound to sites A or B. QM/MM calculations show that there would be an energy barrier of ~15-20 kcal/mol associated with the displacement of the ion from A or B to C. Therefore, if an ion initially binds to A or B it would be probably trapped there instead of moving to C.
Supplementary Figure 5 Accumulation of the nicked product in the K120M mutant.
A) The K120M mutation disturbs double strand cleavage allowing the accumulation of nicked plasmid. 2 nM of supercoiled plasmid DNA target was incubated with 100 nM of wild type I-DmoI or 50, 100 nM of I-DmoI K120M mutant at pH 8, 65 ºC for 30 min. B) Nicked oligonucleotides labeled with 6-FAM (C and NC) were used to determine which DNA strand is affected by the K120M mutation in I-DmoI. Oligonucleotide and protein at 750 nM were incubated at pH 8, 65 ºC for 30 min. The left panel of the gel displays NC and C substrates in the absence of enzyme determining the non-cleaved status of the probes. In the right panel of the gel NC and C were incubated with I-DmoI K120M showing that the mutant activity to digest the intact coding strand in target C is normal; however, the mutant activity was affected when the NC probe (containing the intact non-coding strand) was used in the assay.
Supplementary Figure 6 Potential energy surface at the QM(B3LYP/SVP) level, considering a direct nucleophilic attack of a hydroxyl ion on the target phosphate group.
The same model depicted in Supplementary Figure 7A was considered [RC: Reaction Coordinate (combination of interatomic distances considered to model the reaction of interest)]. After optimization of the initial structure, a penta-coordinated phosphorous intermediate (similar to the one obtained in first step of the mechanism depicted in Supplementary Figure 7) is obtained, later this intermediate easily evolves to the final hydrolytic products (reaction energy of -32.7 kcal/mol at the QM(B3LYP/6-311++G(d,p)//B3LYP/SVP) level of theory). However, the distance between the O3’ leaving atom and the phosphate group as well as the pattern of coordination of metals in sites B and C are inconsistent with the crystallographic structures corresponding to states 5 to 7 of the catalytic mechanism of I-DmoI.
Supplementary Figure 7 QM(B3LYP/SVP) calculations in a model of the I-DmoI active site, considering a proton transfer between the nucleophilic water molecule and the O3′ leaving group.
A) The model considered (110 atoms) was based on the structure corresponding to time 8h (state 3). B) Potential Energy Surface (PES) of a first step where the target phosphate group deprotonates the incoming water molecule and renders a penta-coordinated phosphorous intermediate. The associated energy barrier and reaction energy of this step at the QM(B3LYP/6-311++G(d,p)//B3LYP/SVP) level are 24.8 and 20.4 kcal/mol respectively. C) PES corresponding to the final nucleophilic attack of the resulting hydroxyl ion and the protonation of the O3’ leaving group. The reaction energy considering the final product and the reactants is 1.0 kcal/mol at the QM(B3LYP/6-311++G(d,p)//B3LYP/SVP) level of theory RC: Reaction Coordinate (combination of interatomic distances considered to model the reaction of interest). D) Superimposition of the structure corresponding to the last stage of the catalytic process (state 7) after replacing the water in site C by Mg2+ and the same structure optimized at level QM(B3LYP/SVP)/MM(AMBER). The pre-optimized structure is depicted in dark gray. Notice that E117 is not coordinating the ion in C in the optimized structure, which negatively impacts its binding affinity.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1 and 2 (PDF 1503 kb)
Conformational changes in the active center of I-DmoI
Detailed view of the conformational changes in the active site of I-DmoI during catalysis showing the 7 reactant states. The differences in metal occupancy during the course of the reaction are represented with different tones of grey. The color code is the same as in the main figures 1-4. (MOV 3587 kb)
Detail of the metal exit in site C
View of the E117 conformational change promoting metal exit in site C. The differences in metal occupancy during the course of the reaction are represented with different tones of grey. The color code is the same as in the main figures 1-4. (MOV 1211 kb)
Rights and permissions
About this article
Cite this article
Molina, R., Stella, S., Redondo, P. et al. Visualizing phosphodiester-bond hydrolysis by an endonuclease. Nat Struct Mol Biol 22, 65–72 (2015). https://doi.org/10.1038/nsmb.2932
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2932
This article is cited by
-
DNA cleavage by endonuclease I-DmoI: a QM/MM study and comparison with experimental data provide indications on the environmental effects
Theoretical Chemistry Accounts (2020)
-
Intrinsic cleavage of RNA polymerase II adopts a nucleobase-independent mechanism assisted by transcript phosphate
Nature Catalysis (2019)
-
Understanding the indirect DNA read-out specificity of I-CreI Meganuclease
Scientific Reports (2018)
-
Dynamic coordination of two-metal-ions orchestrates λ-exonuclease catalysis
Nature Communications (2018)
-
Phosphate steering by Flap Endonuclease 1 promotes 5′-flap specificity and incision to prevent genome instability
Nature Communications (2017)