Abstract
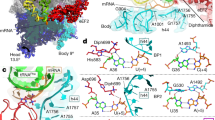
During translation, elongation factor G (EF-G) catalyzes the translocation of tRNA2–mRNA inside the ribosome. Translocation is coupled to a cycle of conformational rearrangements of the ribosomal machinery, and how EF-G initiates translocation remains unresolved. Here we performed systematic mutagenesis of Escherichia coli EF-G and analyzed inhibitory single-site mutants of EF-G that preserved pretranslocation (Pre)-state ribosomes with tRNAs in A/P and P/E sites (Pre–EF-G). Our results suggest that the interactions between the decoding center and the codon–anticodon duplex constitute the barrier for translocation. Catalysis of translocation by EF-G involves the factor's highly conserved loops I and II at the tip of domain IV, which disrupt the hydrogen bonds between the decoding center and the duplex to release the latter, hence inducing subsequent translocation events, namely 30S head swiveling and tRNA2–mRNA movement on the 30S subunit.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Moore, P.B. & Steitz, T.A. The roles of RNA in the synthesis of protein. Cold Spring Harb. Perspect. Biol. 3, a003780 (2011).
Ramakrishnan, V. The ribosome: some hard facts about its structure and hot air about its evolution. Cold Spring Harb. Symp. Quant. Biol. 74, 25–33 (2009).
Frank, J. Intermediate states during mRNA-tRNA translocation. Curr. Opin. Struct. Biol. 22, 778–785 (2012).
Katunin, V.I., Savelsbergh, A., Rodnina, M.V. & Wintermeyer, W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry 41, 12806–12812 (2002).
Frank, J., Gao, H., Sengupta, J., Gao, N. & Taylor, D.J. The process of mRNA-tRNA translocation. Proc. Natl. Acad. Sci. USA 104, 19671–19678 (2007).
Gao, Y.G. et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–699 (2009).
Ratje, A.H. et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468, 713–716 (2010).
Li, W., Trabuco, L.G., Schulten, K. & Frank, J. Molecular dynamics of EF-G during translocation. Proteins 79, 1478–1486 (2011).
Tourigny, D.S., Fernandez, I.S., Kelley, A.C. & Ramakrishnan, V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 340, 1235490 (2013).
Zhou, J., Lancaster, L., Donohue, J.P. & Noller, H.F. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science 340, 1236086 (2013).
Pulk, A. & Cate, J.H. Control of ribosomal subunit rotation by elongation factor G. Science 340, 1235970 (2013).
Chen, Y., Feng, S., Kumar, V., Ero, R. & Gao, Y.G. Structure of EF-G–ribosome complex in a pretranslocation state. Nat. Struct. Mol. Biol. 20, 1077–1084 (2013).
Qin, Y. et al. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 127, 721–733 (2006).
Liu, H., Pan, D., Pech, M. & Cooperman, B.S. Interrupted catalysis: the EF4 (LepA) effect on back-translocation. J. Mol. Biol. 396, 1043–1052 (2010).
Liu, H. et al. The conserved protein EF4 (LepA) modulates the elongation cycle of protein synthesis. Proc. Natl. Acad. Sci. USA 108, 16223–16228 (2011).
Zhang, D. & Qin, Y. The paradox of elongation factor 4: highly conserved, yet of no physiological significance? Biochem. J. 452, 173–181 (2013).
Martemyanov, K.A. & Gudkov, A.T. Domain IV of elongation factor G from Thermus thermophilus is strictly required for translocation. FEBS Lett. 452, 155–159 (1999).
Savelsbergh, A., Matassova, N.B., Rodnina, M.V. & Wintermeyer, W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J. Mol. Biol. 300, 951–961 (2000).
Ogle, J.M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001).
Demeshkina, N., Jenner, L., Westhof, E., Yusupov, M. & Yusupova, G. A new understanding of the decoding principle on the ribosome. Nature 484, 256–259 (2012).
Frank, J. & Agrawal, R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–322 (2000).
Munro, J.B., Sanbonmatsu, K.Y., Spahn, C.M. & Blanchard, S.C. Navigating the ribosome's metastable energy landscape. Trends Biochem. Sci. 34, 390–400 (2009).
Chen, C. et al. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol. Cell 42, 367–377 (2011).
Feng, S., Chen, Y. & Gao, Y.G. Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex. PLoS ONE 8, e58829 (2013).
Yamamoto, H. et al. EF-G and EF4: translocation and back-translocation on the bacterial ribosome. Nat. Rev. Microbiol. 12, 89–100 (2014).
Tsuboi, M. et al. EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis. Mol. Cell 35, 502–510 (2009).
Spahn, C.M. et al. Localization of the ribosomal protection protein Tet(O) on the ribosome and the mechanism of tetracycline resistance. Mol. Cell 7, 1037–1045 (2001).
Connell, S.R. et al. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J. 22, 945–953 (2003).
Dönhöfer, A. et al. Structural basis for TetM-mediated tetracycline resistance. Proc. Natl. Acad. Sci. USA 109, 16900–16905 (2012).
Li, W. et al. Mechanism of tetracycline resistance by ribosomal protection protein Tet(O). Nat. Commun. 4, 1477 (2013).
Gausing, K. Regulation of ribosome production in Escherichia coli: synthesis and stability of ribosomal RNA and of ribosomal protein messenger RNA at different growth rates. J. Mol. Biol. 115, 335–354 (1977).
Connell, S.R. et al. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat. Struct. Mol. Biol. 15, 910–915 (2008).
Czworkowski, J., Wang, J., Seitz, T.A. & Moore, P.B. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J. 13, 3661–3668 (1994).
Moazed, D. & Noller, H.F. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in the 16S rRNA. J. Mol. Biol. 211, 135–145 (1990).
Spiegel, P.C., Ermolenko, D.N. & Noller, H.F. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA 13, 1473–1482 (2007).
Ramrath, D.J. et al. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc. Natl. Acad. Sci. USA 110, 20964–20969 (2013).
Brilot, A.F., Korostelev, A.A., Ermolenko, D.N. & Grigorieff, N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc. Natl. Acad. Sci. USA 110, 20994–20999 (2013).
Matassova, A.B., Rodnina, M.V. & Wintermeyer, W. Elongation factor G-induced structural change in helix 34 of 16S rRNA related to translocation on the ribosome. RNA 7, 1879–1885 (2001).
Zhang, W., Dunkle, J.A. & Cate, J.H. Structures of the ribosome in intermediate states of ratcheting. Science 325, 1014–1017 (2009).
Guo, Z. & Noller, H.F. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc. Natl. Acad. Sci. USA 109, 20391–20394 (2012).
Khade, P.K., Shi, X. & Joseph, S. Steric complementarity in the decoding center is important for tRNA selection by the ribosome. J. Mol. Biol. 425, 3778–3789 (2013).
Ogle, J.M., Murphy, F.V., Tarry, M.J. & Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 (2002).
Agirrezabala, X. et al. Structural insights into cognate versus near-cognate discrimination during decoding. EMBO J. 30, 1497–1507 (2011).
Joseph, S. & Noller, H.F. EF-G-catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO J. 17, 3478–3483 (1998).
Phelps, S.S., Jerinic, O. & Joseph, S. Universally conserved interactions between the ribosome and the anticodon stem-loop of a site tRNA important for translocation. Mol. Cell 10, 799–807 (2002).
Khade, P.K. & Joseph, S. Messenger RNA interactions in the decoding center control the rate of translocation. Nat. Struct. Mol. Biol. 18, 1300–1302 (2011).
Garcia-Ortega, L., Stephen, J. & Joseph, S. Precise alignment of peptidyl tRNA by the decoding center is essential for EF-G-dependent translocation. Mol. Cell 32, 292–299 (2008).
Abdi, N.M. & Fredrick, K. Contribution of 16S rRNA nucleotides forming the 30S subunit A and P sites to translation in Escherichia coli. RNA 11, 1624–1632 (2005).
Rodnina, M.V., Savelsbergh, A., Katunin, V.I. & Wintermeyer, W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385, 37–41 (1997).
Spirin, A.S. The ribosome as a conceying themal ratchet machine. J. Biol. Chem. 284, 21103–21119 (2009).
Zhang, D. et al. Common chaperone activity in the G-domain of trGTPase protects L11-L12 interaction on the ribosome. Nucleic Acids Res. 40, 10851–10865 (2012).
Walker, S.E., Shoji, S., Pan, D., Cooperman, B.S. & Fredrick, K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc. Natl. Acad. Sci. USA 105, 9192–9197 (2008).
Wang, L. et al. A conserved proline switch on the ribosome facilitates the recruitment and binding of trGTPases. Nat. Struct. Mol. Biol. 19, 403–410 (2012).
Hansson, S., Singh, R., Gudkov, A.T., Liljas, A. & Logan, D.T. Structural insights into fusidic acid resistance and sensitivity in EF-G. J. Mol. Biol. 348, 939–949 (2005).
Martemyanov, K.A., Liljas, A. & Gudkov, A.T. Extremely thermostable elongation factor G from Aquifex aeolicus: cloning, expression, purification, and characterization in a heterologous translation system. Protein Expr. Purif. 18, 257–261 (2000).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Bailey, C.H. & Kandel, E.R. Structural changes accompanying memory storage. Annu. Rev. Physiol. 55, 397–426 (1993).
Laurberg, M. et al. Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J. Mol. Biol. 303, 593–603 (2000).
Youngman, E.M., Brunelle, J.L., Kochaniak, A.B. & Green, R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117, 589–599 (2004).
Borowski, C., Rodnina, M.V. & Wintermeyer, W. Truncated elongation factor G lacking the G domain promotes translocation of the 3′ end but not of the anticodon domain of peptidyl-tRNA. Proc. Natl. Acad. Sci. USA 93, 4202–4206 (1996).
Hickerson, R., Majumdar, Z.K., Baucom, A., Clegg, R.M. & Noller, H.F. Measurement of internal movements within the 30 S ribosomal subunit using Forster resonance energy transfer. J. Mol. Biol. 354, 459–472 (2005).
Culver, G.M. & Noller, H.F. In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol. 318, 446–460 (2000).
Lyu, Z.X., Shao, Q., Gao, Y.Q. & Zhao, X.S. Direct observation of the uptake of outer membrane proteins by the periplasmic chaperone Skp. PLoS ONE 7, e46068 (2012).
Acknowledgements
We thank members of the P. Zhu laboratory and N. Gao for help and discussion. Y.Q. is supported by the Institute of Biophysics 135 Goal-oriented project, National Laboratory of Biomacromolecules (Institute of Biophysics, Chinese Academy of Sciences), and the State Key Laboratory of Molecular Biology (Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences). This work was supported by grants from the Ministry of Science and Technology of China (2012CB911000 and 2013CB531200 to Y.Q.), the National Natural Science Foundation of China (31322015, 31170756 and 31270847 to Y.Q.) and the Chinese Academy of Sciences (project KSZD-EW-Z-003 to Y.Q.).
Author information
Authors and Affiliations
Contributions
G.L., G.S., Danyang Zhang, Dejiu Zhang and Z. Li cloned constructs and performed biochemical assays. Z. Lyu and G.S. collected FRET data and with X.S.Z. analyzed the FRET data. J.D. collected X-ray data and with W.G. resolved the structures. J.A. prepared some figures and analyzed data. Y.Q. and K.H.N. analyzed all data and wrote the manuscript. All authors discussed the results and commented on the manuscript. Y.Q. directed and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
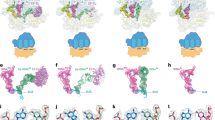
Supplementary Figure 1 Sequence and structure alignment of EF-G homologs.
(a) Sequence alignments of loops I and II of EF-G and homologues. EF-G, bacterial EF-G; mtEF-G1, mitochondrial EF-G orthologue (translocase); mtEF-G2, mitochondrial paralogue (terminase); Tet(O), members of the Tet(O) family mediating tetracycline resistance. Red, 100% identity of the amino acid sequence with the bacterial consensus sequence of EF-G; blue, conservative amino acid substitution.(b) The overall structure alignment of Tet (O)–GTP conformer on the ribosome (orange, PDB 3J36) in comparison with EF-G–GTP in EF-G–POST (cyan, PDB 2WRI). Domains I-III and V are indicated as common domains. The dashed line surrounded region is the specific domain, i.e. domain IV (D IV). D IVs from EF-G (dark blue) and Tet(O) (dark yellow) are aligned and zoomed-in with 90° turn.
Supplementary Figure 2 In vivo experiments of EF-G and the mutants.
(a) Colonies of E. coli cells over-expressing EF-G or its mutants on agar plates. (b) Polysome pattern of the cells in a. Without IPTG treatment (IPTG-), all mutants exhibited similar polysomes pattern as in WT EF-G cell (black curves). When the cells have been induced with ITPG, the polysomes patterns showing difference are indicated in red (strong) and pink (mild).
Supplementary Figure 3 Binding assays of EF-G and its mutants.
(a) Binding of EF-G or its mutants to vacant ribosomes was analyzed by sucrose cushion ultracentrifugation and SDS-PAGE. Bands corresponding to EF-G and the ribosomal protein S1 are indicated by the arrows. Pre: reaction mix before sucrose cushion. Su/Pe: Supernatant/Pellet after sucrose cushion. (b) The quantification of chemical probing results. TO I is shown as an example (red rectangle). Error bars, s.e.m. (n = 3 technical replicates). **P<0.01, ***P<0.001 by two-tailed Student's t test.
Supplementary Figure 4 FRET analyses of 30S head swiveling.
30S head swiveling indicated by changes in FRET efficiencies between donor Alexa 555 and acceptor Alexa 647 probes attached to S11 and S13 for the S11-Donor/S13-Acceptor pair. PRE 70S complexes were rapidly mixed with EF-G wt or the EF-G mutant S588P or Δloop II and GTP at time 0 and Alexa 647 emission was recorded. Each trace represents the average of 10 reactions.
Supplementary Figure 5 Structural comparison EF-G and D IV loops in solution and on the ribosome.
(a) The overall structure alignment of EF-G in the GTP (PDB 2OM7) and GDP (PDB 2WRI) conformations on the ribosome. When D IVs are aligned and zoomed-in with 90° turn, the tip of loop I moved 10 Å towards P-site upon GTP hydrolysis, whereas loop II remained unchanged. (b) The same proteins of a but in solution. GTP conformer (PDB 1WDT), GDP conformer (PDB 1FNM). The upper tip of D V moved 25 Å. When D IVs were aligned and zoomed-in, the tip of loop I moved 8 Å in the opposite direction compared to its conformation on the ribosome. (c) Structural comparison of EF-G on POST complexes. ecEF-G and ttEF-G are extracted from E. coli and T. thermuphilus POST–EF-G complex (PDB 4KIY and 4KBV), respectively. The overall structures of the two EF-Gs were aligned or D IVs were aligned and zoomed-in with 90° turn. (d) The interface of EF-G with the DC in EF-G–POST (PDB 2WRI). Common domains of EF-G contact only the ribosome whereas D IV interacts directly with tRNA2–mRNA through its tip region. (e) The zoom-in view of the interface between D IV tip with P-tRNA–mRNA or with DC. A-tRNA (yellow) has been extracted from EF-Tu–PRE (PDB 2Y0U).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–3 (PDF 8973 kb)
Rights and permissions
About this article
Cite this article
Liu, G., Song, G., Zhang, D. et al. EF-G catalyzes tRNA translocation by disrupting interactions between decoding center and codon–anticodon duplex. Nat Struct Mol Biol 21, 817–824 (2014). https://doi.org/10.1038/nsmb.2869
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2869
This article is cited by
-
Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation
Nature Communications (2021)
-
Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP
Nature Communications (2021)
-
Insights into genome recoding from the mechanism of a classic +1-frameshifting tRNA
Nature Communications (2021)
-
The ribosome moves: RNA mechanics and translocation
Nature Structural & Molecular Biology (2017)
-
EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nature Structural & Molecular Biology (2016)