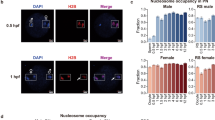
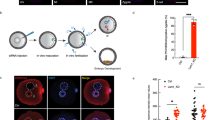
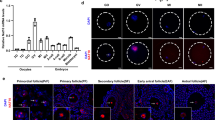
Abstract
Packaging of DNA into nucleosomes not only helps to store genetic information but also creates diverse means for regulating DNA-templated processes. Attempts to reveal additional functions of the nucleosome have been unsuccessful, owing to cell lethality caused by nucleosome deletion. Taking advantage of the mammalian fertilization process, in which sperm DNA assembles into nucleosomes de novo, we generated nucleosome-depleted (ND) paternal pronuclei by depleting maternal histone H3.3 or its chaperone HIRA in mouse zygotes. We found that the ND pronucleus forms a nuclear envelope devoid of nuclear pore complexes (NPCs). Loss of NPCs is accompanied by defective localization of ELYS, a nucleoporin essential for NPC assembly, to the nuclear rim. Interestingly, tethering ELYS to the nuclear rim of the ND nucleus rescues NPC assembly. Our study thus demonstrates that nucleosome assembly is a prerequisite for NPC assembly during paternal pronuclear formation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Kornberg, R.D. & Lorch, Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 (1999).
Luger, K., Dechassa, M. & Tremethick, D. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 13, 436–447 (2012).
Burgess, R.J. & Zhang, Z. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 20, 14–22 (2013).
Margueron, R. & Reinberg, D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 11, 285–296 (2010).
Marzluff, W.F., Gongidi, P., Woods, K., Jin, J. & Maltais, L. The human and mouse replication-dependent histone genes. Genomics 80, 487–498 (2002).
Rykowski, M.C., Wallis, J., Choe, J. & Grunstein, M. Histone H2B subtypes are dispensable during the yeast cell cycle. Cell 25, 477–487 (1981).
Drané, P., Ouararhni, K., Depaux, A., Shuaib, M. & Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 (2010).
Goldberg, A.D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010).
Ray-Gallet, D. et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 (2011).
Jenkins, T.G. & Carrell, D. Dynamic alterations in the paternal epigenetic landscape following fertilization. Front. Genet. 3, 143 (2012).
Nonchev, S. & Tsanev, R. Protamine-histone replacement and DNA replication in the male mouse pronucleus. Mol. Reprod. Dev. 25, 72–76 (1990).
van der Heijden, G.W. et al. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 122, 1008–1022 (2005).
Elsaesser, S.J., Goldberg, A.D. & Allis, C.D. New functions for an old variant: no substitute for histone H3.3. Curr. Opin. Genet. Dev. 20, 110–117 (2010).
Talbert, P.B. & Henikoff, S. Histone variants: ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275 (2010).
Akiyama, T., Suzuki, O., Matsuda, J. & Aoki, F. Dynamic replacement of histone h3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet. 7, e1002279 (2011).
Torres-Padilla, M.E., Bannister, A., Hurd, P., Kouzarides, T. & Zernicka-Goetz, M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50, 455–461 (2006).
Loppin, B. et al. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437, 1386–1390 (2005).
Inoue, A. & Aoki, F. Role of the nucleoplasmin 2 C-terminal domain in the formation of nucleolus-like bodies in mouse oocytes. FASEB J. 24, 485–494 (2010).
Smith, S. & Stillman, B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10, 971–980 (1991).
Nashun, B., Yukawa, M., Liu, H., Akiyama, T. & Aoki, F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development 137, 3785–3794 (2010).
D'Angelo, M.A., Anderson, D., Richard, E. & Hetzer, M. Nuclear pores form de novo from both sides of the nuclear envelope. Science 312, 440–443 (2006).
Webster, M., Witkin, K. & Cohen-Fix, O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 122, 1477–1486 (2009).
Güttinger, S., Laurell, E. & Kutay, U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10, 178–191 (2009).
Newport, J.W., Wilson, K. & Dunphy, W. A lamin-independent pathway for nuclear envelope assembly. J. Cell Biol. 111, 2247–2259 (1990).
Gu, T.P. et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011).
Clarke, P.R. & Zhang, C. Ran GTPase: a master regulator of nuclear structure and function during the eukaryotic cell division cycle? Trends Cell Biol. 11, 366–371 (2001).
Quimby, B.B. & Dasso, M. The small GTPase Ran: interpreting the signs. Curr. Opin. Cell Biol. 15, 338–344 (2003).
Doucet, C.M., Talamas, J. & Hetzer, M. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 141, 1030–1041 (2010).
Franz, C. et al. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 8, 165–172 (2007).
Gillespie, P.J., Khoudoli, G., Stewart, G., Swedlow, J. & Blow, J. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr. Biol. 17, 1657–1662 (2007).
Galy, V., Askjaer, P., Franz, C., López-Iglesias, C. & Mattaj, I. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr. Biol. 16, 1748–1756 (2006).
Bilokapic, S. & Schwartz, T. Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains. Structure 21, 572–580 (2013).
Rasala, B.A., Ramos, C., Harel, A. & Forbes, D. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol. Biol. Cell 19, 3982–3996 (2008).
Lau, C.K. et al. Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly. Mol. Biol. Cell 20, 4043–4058 (2009).
Fernandez, A.G. & Piano, F. MEL-28 is downstream of the Ran cycle and is required for nuclear-envelope function and chromatin maintenance. Curr. Biol. 16, 1757–1763 (2006).
Dumont, J. et al. A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J. Cell Biol. 176, 295–305 (2007).
Hughes, M., Zhang, C., Avis, J., Hutchison, C. & Clarke, P. The role of the ran GTPase in nuclear assembly and DNA replication: characterisation of the effects of Ran mutants. J. Cell Sci. 111, 3017–3026 (1998).
Inoue, A., Ogushi, S., Saitou, M., Suzuki, M. & Aoki, F. Involvement of mouse nucleoplasmin 2 in the decondensation of sperm chromatin after fertilization. Biol. Reprod. 85, 70–77 (2011).
Bonnefoy, E., Orsi, G., Couble, P. & Loppin, B. The essential role of Drosophila HIRA for de novo assembly of paternal chromatin at fertilization. PLoS Genet. 3, 1991–2006 (2007).
Philpott, A. & Leno, G.H. Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell 69, 759–767 (1992).
Forbes, D.J., Kirschner, M. & Newport, J. Spontaneous formation of nucleus-like structures around bacteriophage DNA microinjected into Xenopus eggs. Cell 34, 13–23 (1983).
Newport, J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell 48, 205–217 (1987).
Ulbert, S., Platani, M., Boue, S. & Mattaj, I. Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J. Cell Biol. 173, 469–476 (2006).
Anderson, D.J. & Hetzer, M. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat. Cell Biol. 9, 1160–1166 (2007).
Szymborska, A. et al. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341, 655–658 (2013).
D'Angelo, M.A. & Hetzer, M.W. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18, 456–466 (2008).
Rotem, A. et al. Importin β regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol. Biol. Cell 20, 4031–4042 (2009).
Imamoto, N. & Funakoshi, T. Nuclear pore dynamics during the cell cycle. Curr. Opin. Cell Biol. 24, 453–459 (2012).
Malik, H.S. & Henikoff, S. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10, 882–891 (2003).
Pesty, A., Miyara, F., Debey, P., Lefevre, B. & Poirot, C. Multiparameter assessment of mouse oogenesis during follicular growth in vitro. Mol. Hum. Reprod. 13, 3–9 (2007).
Hirao, Y. Isolation of ovarian components essential for growth and development of mammalian oocytes in vitro. J. Reprod. Dev. 58, 167–174 (2012).
Yamaguchi, S. et al. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 23, 329–339 (2013).
Ooga, M. et al. Changes in H3K79 methylation during preimplantation development in mice. Biol. Reprod. 78, 413–424 (2008).
Okada, Y., Yamagata, K., Hong, K., Wakayama, T. & Zhang, Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature 463, 554–558 (2010).
Ellenberg, J. et al. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138, 1193–1206 (1997).
Clever, M., Funakoshi, T., Mimura, Y., Takagi, M. & Imamoto, N. The nucleoporin ELYS/Mel28 regulates nuclear envelope subdomain formation in HeLa cells. Nucleus 3, 187–199 (2012).
Fang, J., Wang, H. & Zhang, Y. Purification of histone methyltransferases from HeLa cells. Methods Enzymol. 377, 213–226 (2004).
Acknowledgements
We thank T. Haraguchi (Osaka University) and T. Kirchhausen (Harvard Medical School) for helpful discussion, F. Aoki (University of Tokyo) for the EGFP-NLS, Flag-H2B and Flag-H4 constructs, and I. Mattaj (European Molecular Biology Laboratory) and G.L. Xu (Chinese Academy of Sciences) for antibodies. We thank T. Haraguchi, G. German, S. Yamaguchi, S. Matoba and D. Cai for critical reading of the manuscript. This work was supported by US National Institutes of Health (U01DK089565 to Y.Z.). A.I. is supported as a research fellow for Research Abroad of the Japan Society for the Promotion of Science. Y.Z. is supported as an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
A.I. and Y.Z. conceived the project, designed the experiments and wrote the manuscript; A.I. performed experiments and analyzed the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 siRNA-mediated knockdown of maternal HIRA protein.
(a) Experimental scheme for siRNA-mediated knockdown. GV oocytes were injected with siRNA and cultured for 18 h to reach the MII-stage. Following meiotic maturation, oocytes were fertilized in vitro. (b) RT-qPCR analysis of the Hira mRNA levels in MII oocytes. The relative expression was normalized to GAPDH. The value of Control was set as 1. (c) Representative images of fertilized oocytes stained with anti-HIRA (green) antibody and DAPI (blue) at 2 hrs after fertilization. P; paternal chromatin. M; maternal chromatin. Scale bar, 20 μm. (d) Experimental scheme for siRNA-mediated knockdown of HIRA using the in vitro growth system. Growing oocytes surrounded by granulosa layers are isolated from 12 days old female mice, injected with siRNA, and cultured for 12 days in vitro until they reach the fully grown stage. Following in vitro growth, oocytes are matured and fertilized in vitro. (e) Microinjection into growing oocytes surrounded by granulosa cells. Scale bar, 50 μm. (f) Representative images of oocyte-granulosa cell complex during oocytes growth in vitro of the indicated times. Scale bar, 200 μm. (g) In vitro maturation of cumulus-oocyte complexes following IVG. Cumulus cells surrounding oocytes are expanded during meiotic maturation. Scale bars, 100 μm.
Supplementary Figure 2 Protamine removal is independent of histone deposition in mouse zygotes.
(a) Evaluation of anti-protamine 2 (Pro2) antibody. Surface spread samples of spermatozoa were stained with Pro2 antibody (green) and DAPI (blue). As a negative control, the sample was treated with the secondary antibody only. Scale bar, 10 μm. (b) Surface spread samples of fertilized oocytes were stained with Pro2 (green) and anti-H2AX (red) antibodies and DAPI (blue) at 1 h or 1.5 h post-fertilization (hpf). Note that protamines are still detectable in the condensed region in which H2AX has not been incorporated at 1 h. At 1.5 h, protamines are undetectable in the decondensed paternal DNA in which H2AX is broadly incorporated. A spermatozoon (Sp), which did not enter the oocyte, was used as a positive control for Pro2 staining. Scale bar, 10 μm. (c, d) Representative images of fertilized oocytes stained with Pro2 antibody (white) and DAPI (blue) at 2 hpf. Whole mount samples (c) and surface spread samples (d) were prepared. Note that protamines are undetectable in the paternal DNA of both control and HIRA-depleted zygotes (pointed by arrows). Dotted lines represent the paternal DNA entering the oocytes. The total numbers of zygotes examined were 10-14 and 8-10 per group for the whole mount and surface spread, respectively. Sp; spermatozoa that do not enter the oocyte. P; paternal DNA. M; maternal DNA. PB; polar body. Scale bars, 20 μm.
Supplementary Figure 3 Expression of histone genes and localization of HIRA in H3.3-depleted zygotes.
(a) RT-qPCR analysis of oocytes injected with H3.3 siRNA. GV oocytes were injected with a mixture of siRNAs against H3.3a and H3.3b or control siRNA. Twenty-four hours after microinjection, the oocytes were matured in vitro to reach the MII-stage. RNA was extracted from the MII oocytes and the expression levels of indicated genes were analyzed by RT-qPCR. The relative expression level was normalized to Gapdh. The values of control oocytes were set as 1. The experiment was repeated twice. Error bars indicate as s.d. (b) Representative images of fertilized oocytes stained with anti-HIRA (green), anti-H2AX (red), and DAPI (blue) at 2 hours after fertilization. Note that HIRA localizes to the paternal DNA, where H2AX is not detected, in H3.3-depleted zygotes. A total of 25 zygotes per group were examined. Percentages indicate zygotes showing the depicted phenotype. P; paternal chromatin. M; maternal chromatin. PB; polar body. Scale bar, 20 μm.
Supplementary Figure 4 TEM images of pronuclei in zygotes.
Shown are representative images of the paternal and maternal pronucleus in control or H3.3-depleted zygotes. Asterisks indicate nucleolus-like bodies. Scale bars, 2.5 μm.
Supplementary Figure 5 Dynamics of ELYS during maternal pronuclear formation.
(a, b) Representative images of the maternal chromatin in control (a) and H3.3-depleted (b) zygotes stained with mAb414 (blue) and DAPI at the indicated times of post-fertilization (hpf). EGFP-ELYS (green) and LBR-RFP (red) mRNAs were injected at GV-stage. Merge = green + red channels. A total of 4-7 and 10-12 of control and H3.3-depleted zygotes, respectively, per time point were examined. Scale bars, 10 μm. (c) Surface spread staining of control zygotes stained with anti-H2AX (red) and Crest (white; centromere marker) antibodies at 2 hpf. The right panel indicates enlarged images of maternal chromosomes, showing that EGFP-ELYS is localized in the rim of maternal chromosome mass.
Supplementary Figure 6 Neither Nup37 nor POM121 is localized to the rim of nucleosome-depleted paternal pronucleus.
(a) Representative images of zygotes stained with mAb414 (Magenta) and DAPI at 8 hrs post-fertilization (hpf). Nup37-EGFP (green) mRNA was injected at GV-stage. A total of 13 or 26 of control or H3.3-depleted zygotes, respectively, were examined. Percentages indicate zygotes showing the depicted phenotype. (b) Representative images of zygotes stained with anti-POM121 (Magenta) and mAb414 (green) antibodies and DAPI at 8 hpf. A total of 7 or 13 of control or H3.3-depleted zygotes, respectively, were examined. P; paternal pronucleus (pointed by arrows). M; maternal pronucleus. Scale bars, 20 μm.
Supplementary Figure 8 Loss of Ran-mediated protein transport does not prevent the localization of ELYS to nuclear rim in zygotes.
(a) Western blot analysis showing the expression of wild-type and Ran mutants at similar levels. GV oocytes were injected with RanWT-mRFP1 (monomeric red fluorescent protein 1), RanT24N–mRFP1, or RanQ69L–mRFP1 mRNA. Following meiotic maturation, extracts of MII oocytes were subjected to Western blot analysis. Tubulin was used as a loading control. Molecular weight (kDa) is shown on the left. Shown is a representative result from two biologically independent experiments. (b) Representative image of the paternal pronucleus in zygotes stained with anti-lamin B1 antibody at 8 hrs post-fertilization (hpf). A total of 7, 10, and 9 zygotes in WT, T24N, and Q69L, respectively, were examined. Percentages indicate zygotes showing the depicted phenotype. Scale bar, 20 μm. (c) Representative images of the paternal and maternal chromatin in zygotes stained with anti-H2AX antibody at 2 hpf. EGFP-ELYS mRNA was co-injected with each form of Ran mRNA. A total of 12, 11, and 11 zygotes in WT, T24N, and Q69L, respectively, were examined. Scale bar, 10 μm. (d) Representative images of EGFP-ELYS and NPC (marked by the mAb414) in the paternal and maternal pronucleus of zygotes at 8 hpf. A total of 15, 20, and 13 zygotes in WT, T24N, and Q69L, respectively, were examined. Scale bar, 10 μm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Note (PDF 35928 kb)
Rights and permissions
About this article
Cite this article
Inoue, A., Zhang, Y. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat Struct Mol Biol 21, 609–616 (2014). https://doi.org/10.1038/nsmb.2839
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2839
This article is cited by
-
Actin-driven chromosome clustering facilitates fast and complete chromosome capture in mammalian oocytes
Nature Cell Biology (2023)
-
B-type Plexins promote the GTPase activity of Ran to affect androgen receptor nuclear translocation in prostate cancer
Cancer Gene Therapy (2023)
-
Parental competition for the regulators of chromatin dynamics in mouse zygotes
Communications Biology (2022)
-
H3.3 kinetics predicts chromatin compaction status of parental genomes in early embryos
Reproductive Biology and Endocrinology (2021)
-
Symmetrically dimethylated histone H3R2 promotes global transcription during minor zygotic genome activation in mouse pronuclei
Scientific Reports (2021)