Abstract
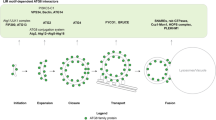
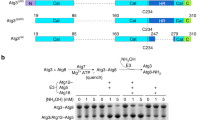
Two autophagy-related ubiquitin-like systems have unique features: the E2 enzyme Atg3 conjugates the ubiquitin-like protein Atg8 to the lipid phosphatidylethanolamine, and the other ubiquitin-like protein conjugate Atg12–Atg5 promotes that conjugase activity of Atg3. Here, we elucidate the mode of this action of Atg12–Atg5 as a new E3 enzyme by using Saccharomyces cerevisiae proteins. Biochemical analyses based on structural information suggest that Atg3 requires a threonine residue to catalyze the conjugation reaction instead of the typical asparagine residue used by other E2 enzymes. Moreover, the catalytic cysteine residue of Atg3 is arranged in the catalytic center such that the conjugase activity is suppressed; Atg12–Atg5 induces a reorientation of the cysteine residue toward the threonine residue, which enhances the conjugase activity of Atg3. Thus, this study reveals the mechanism of the key reaction that drives membrane biogenesis during autophagy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
Mizushima, N., Levine, B., Cuervo, A.M. & Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008).
Nakatogawa, H., Suzuki, K., Kamada, Y. & Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 (2009).
Chen, Y. & Klionsky, D.J. The regulation of autophagy – unanswered questions. J. Cell Sci. 124, 161–170 (2011).
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 (2011).
Rubinsztein, D.C., Shpilka, T. & Elazar, Z. Mechanisms of autophagosome biogenesis. Curr. Biol. 22, R29–R34 (2012).
Yang, Z. & Klionsky, D.J. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814–822 (2010).
Kirisako, T. et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276 (2000).
Kim, J., Huang, W.P. & Klionsky, D.J. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 152, 51–64 (2001).
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000).
Kirisako, T. et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446 (1999).
Nakatogawa, H., Ichimura, Y. & Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 (2007).
Xie, Z., Nair, U. & Klionsky, D.J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19, 3290–3298 (2008).
Nakatogawa, H., Ishii, J., Asai, E. & Ohsumi, Y. Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy 8, 177–186 (2012).
Nair, U. et al. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy 8, 780–793 (2012).
Yu, Z.Q. et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 8, 883–892 (2012).
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998).
Hanada, T. et al. The Atg12–Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302 (2007).
Fujioka, Y. et al. In vitro reconstitution of plant ATG8 and ATG12 conjugation systems essential for autophagy. J. Biol. Chem. 283, 1921–1928 (2008).
Suzuki, N.N., Yoshimoto, K., Fujioka, Y., Ohsumi, Y. & Inagaki, F. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy 1, 119–126 (2005).
Matsushita, M. et al. Structure of Atg5-Atg16, a complex essential for autophagy. J. Biol. Chem. 282, 6763–6772 (2007).
Yamada, Y. et al. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 282, 8036–8043 (2007).
Yamaguchi, M. et al. Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 285, 29599–29607 (2010).
Tong, H., Hateboer, G., Perrakis, A., Bernards, R. & Sixma, T.K. Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system. J. Biol. Chem. 272, 21381–21387 (1997).
Wu, P.Y. et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 22, 5241–5250 (2003).
Yunus, A.A. & Lima, C.D. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 13, 491–499 (2006).
Ichimura, Y. et al. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J. Biol. Chem. 279, 40584–40592 (2004).
Oh-oka, K., Nakatogawa, H. & Ohsumi, Y. Physiological pH and acidic phospholipids contribute to substrate specificity in lipidation of Atg8. J. Biol. Chem. 283, 21847–21852 (2008).
Frand, A.R. & Kaiser, C.A. Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell 4, 469–477 (1999).
Cevc, G. Phospholipids Handbook (Marcel Dekker, 1993).
Suzuki, K. et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981 (2001).
Yamaguchi, M. et al. Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7. Nat. Struct. Mol. Biol. 19, 1250–1256 (2012).
Hanada, T., Satomi, Y., Takao, T. & Ohsumi, Y. The amino-terminal region of Atg3 is essential for association with phosphatidylethanolamine in Atg8 lipidation. FEBS Lett. 583, 1078–1083 (2009).
Zinser, E. & Daum, G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast 11, 493–536 (1995).
Adams, A., Gottschling, D.E., Kaiser, C.A. & Stearns, T. Methods in Yeast Genetics (Cold Spring Harbor Laboratory, 1998).
Brachmann, C.B. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998).
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004).
Yamada, Y. et al. Crystallization and preliminary X-ray analysis of Atg3. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 1016–1017 (2006).
Noda, T., Matsuura, A., Wada, Y. & Ohsumi, Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210, 126–132 (1995).
Hanada, T. & Ohsumi, Y. Structure-function relationship of Atg12, a ubiquitin-like modifier essential for autophagy. Autophagy 1, 110–118 (2005).
Nakatogawa, H. & Ohsumi, Y. SDS-PAGE techniques to study ubiquitin-like conjugation systems in yeast autophagy. Methods Mol. Biol. 832, 519–529 (2012).
Acknowledgements
We thank the members of our laboratories for materials and helpful discussions. This work was supported in part by the Funding Program for Next Generation World-Leading Researchers HO220017 (to H.N.) and Grants-in-Aid for Scientific Research 23000015 (to Y.O.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Contributions
M.S.-N. performed in vitro and in vivo experiments with the help of E.A., H.K. and J.I.; K.M., N.N.N. and F.I. contributed to structural analysis. M.S.-N., H.N. and Y.O. contributed to experimental design and data interpretation and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Note (PDF 3164 kb)
Rights and permissions
About this article
Cite this article
Sakoh-Nakatogawa, M., Matoba, K., Asai, E. et al. Atg12–Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol 20, 433–439 (2013). https://doi.org/10.1038/nsmb.2527
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2527
This article is cited by
-
Multifaceted membrane interactions of human Atg3 promote LC3-phosphatidylethanolamine conjugation during autophagy
Nature Communications (2023)
-
Autophagy-related genes in Egyptian patients with Behçet's disease
Egyptian Journal of Medical Human Genetics (2022)
-
An N-terminal conserved region in human Atg3 couples membrane curvature sensitivity to conjugase activity during autophagy
Nature Communications (2021)
-
TECPR1 promotes aggrephagy by direct recruitment of LC3C autophagosomes to lysosomes
Nature Communications (2020)
-
Role of ABT888, a Novel Poly(ADP-Ribose) Polymerase (PARP) Inhibitor in Countering Autophagy and Apoptotic Processes Associated to Spinal Cord Injury
Molecular Neurobiology (2020)