Abstract
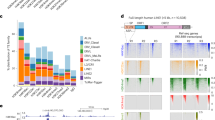
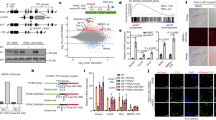
How a more plastic chromatin state is maintained and reversed during development is unknown. Heterochromatin-mediated silencing of repetitive elements occurs in differentiated cells. Here, we used repetitive elements, including retrotransposons, as model loci to address how and when heterochromatin forms during development. RNA sequencing throughout early mouse embryogenesis revealed that repetitive-element expression is dynamic and stage specific, with most repetitive elements becoming repressed before implantation. We show that LINE-1 and IAP retrotransposons become reactivated from both parental genomes after fertilization. Chromatin immunoprecipitation for H3K4me3 and H3K9me3 in 2- and 8-cell embryos indicates that their developmental silencing follows loss of activating marks rather than acquisition of conventional heterochromatic marks. Furthermore, short LINE-1 RNAs regulate LINE-1 transcription in vivo. Our data indicate that reprogramming after mammalian fertilization comprises a robust transcriptional activation of retrotransposons and that repetitive elements are initially regulated through RNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Referenced accessions
NCBI Reference Sequence
References
Waterston, R.H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Waldman, A.S. Ensuring the fidelity of recombination in mammalian chromosomes. Bioessays 30, 1163–1171 (2008).
Maksakova, I.A., Mager, D.L. & Reiss, D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol. Life Sci. 65, 3329–3347 (2008).
Efroni, S. et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437–447 (2008).
Peaston, A.E. et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606 (2004).
Vasseur, M., Condamine, H. & Duprey, P. RNAs containing B2 repeated sequences are transcribed in the early stages of mouse embryogenesis. EMBO J. 4, 1749–1753 (1985).
Svoboda, P. et al. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 269, 276–285 (2004).
Pushkarev, D., Neff, N.F. & Quake, S.R. Single-molecule sequencing of an individual human genome. Nat. Biotechnol. 27, 847–850 (2009).
Pikó, L., Hammons, M.D. & Taylor, K.D. Amounts, synthesis, and some properties of intracisternal A particle-related RNA in early mouse embryos. Proc. Natl. Acad. Sci. USA 81, 488–492 (1984).
Packer, A.I., Manova, K. & Bachvarova, R.F. A discrete LINE-1 transcript in mouse blastocysts. Dev. Biol. 157, 281–283 (1993).
Evsikov, A.V. et al. Systems biology of the 2-cell mouse embryo. Cytogenet. Genome Res. 105, 240–250 (2004).
Bachvarova, R. Small B2 RNAs in mouse oocytes, embryos, and somatic tissues. Dev. Biol. 130, 513–523 (1988).
Plessy, C. et al. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat. Methods 7, 528–534 (2010).
Kramerov, D.A., Bukrinsky, M.I. & Ryskov, A.P. DNA sequences homologous to long double-stranded RNA. Transcription of intracisternal A-particle genes and major long repeat of the mouse genome. Biochim. Biophys. Acta 826, 20–29 (1985).
Rothstein, J.L. et al. Gene expression during preimplantation mouse development. Genes Dev. 6, 1190–1201 (1992).
Watanabe, T. et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543 (2008).
Ohnishi, Y. et al. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Res. 38, 5141–5151 (2010).
Naas, T.P. et al. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 17, 590–597 (1998).
Lunyak, V.V. et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science 317, 248–251 (2007).
Lane, N. et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35, 88–93 (2003).
Smith, Z.D. et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344 (2012).
Taylor, K.D. & Piko, L. Patterns of mRNA prevalence and expression of B1 and B2 transcripts in early mouse embryos. Development 101, 877–892 (1987).
Walsh, C.P., Chaillet, J.R. & Bestor, T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20, 116–117 (1998).
Lehnertz, B. et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192–1200 (2003).
Matsui, T. et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 (2010).
Rowe, H.M. et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 (2010).
Mayer, W., Niveleau, A., Walter, J., Fundele, R. & Haaf, T. Demethylation of the zygotic paternal genome. Nature 403, 501–502 (2000).
Wossidlo, M. et al. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 29, 1877–1888 (2010).
Santos, F., Hendrich, B., Reik, W. & Dean, W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241, 172–182 (2002).
Daujat, S. et al. H3K64 trimethylation marks heterochromatin and is dynamically remodeled during developmental reprogramming. Nat. Struct. Mol. Biol. 16, 777–781 (2009).
Santos, F., Peters, A.H., Otte, A.P., Reik, W. & Dean, W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 280, 225–236 (2005).
Burton, A. & Torres-Padilla, M.E. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief Funct. Genomics 9, 444–454 (2010).
Coufal, N.G. et al. L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131 (2009).
Muotri, A.R. et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910 (2005).
Wiznerowicz, M. et al. The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J. Biol. Chem. 282, 34535–34541 (2007).
Schmitz, K.M., Mayer, C., Postepska, A. & Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24, 2264–2269 (2010).
Giovannangeli, C. & Helene, C. Triplex-forming molecules for modulation of DNA information processing. Curr. Opin. Mol. Ther. 2, 288–296 (2000).
Hoyne, P.R. & Maher, L.J. III. Functional studies of potential intrastrand triplex elements in the Escherichia coli genome. J. Mol. Biol. 318, 373–386 (2002).
Maher, L.J. III. DNA triple-helix formation: an approach to artificial gene repressors? Bioessays 14, 807–815 (1992).
Han, H. & Dervan, P.B. Sequence-specific recognition of double helical RNA and RNA.DNA by triple helix formation. Proc. Natl. Acad. Sci. USA 90, 3806–3810 (1993).
Gorab, E., Amabis, J.M., Stocker, A.J., Drummond, L. & Stollar, B.D. Potential sites of triple-helical nucleic acid formation in chromosomes of Rhynchosciara (Diptera: Sciaridae) and Drosophila melanogaster. Chromosome Res. 17, 821–832 (2009).
Stollar, B.D. & Raso, V. Antibodies recognise specific structures of triple-helical polynucleotides built on poly(A) or poly(dA). Nature 250, 231–234 (1974).
Aravin, A.A., Sachidanandam, R., Girard, A., Fejes-Toth, K. & Hannon, G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747 (2007).
Carmell, M.A. et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514 (2007).
Yang, N. & Kazazian, H.H. Jr. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13, 763–771 (2006).
Faulkner, G.J. et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 41, 563–571 (2009).
Taft, R.J. et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 41, 572–578 (2009).
Chow, J.C. et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141, 956–969 (2010).
Puschendorf, M. et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 40, 411–420 (2008).
Miyanari, Y. & Torres-Padilla, M.E. Control of ground-state pluripotency by allelic regulation of Nanog. Nature 483, 470–473 (2012).
Torres-Padilla, M.E., Bannister, A.J., Hurd, P.J., Kouzarides, T. & Zernicka-Goetz, M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50, 455–461 (2006).
Santenard, A. et al. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol. 12, 853–862 (2010).
Dewannieux, M., Dupressoir, A., Harper, F., Pierron, G. & Heidmann, T. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat. Genet. 36, 534–539 (2004).
Boyer, L.A. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005).
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008).
Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005).
Alunni-Fabbroni, M., Manfioletti, G., Manzini, G. & Xodo, L.E. Inhibition of T7 RNA polymerase transcription by phosphate and phosphorothioate triplex-forming oligonucleotides targeted to a R.Y site downstream from the promoter. Eur. J. Biochem. 226, 831–839 (1994).
Acknowledgements
We thank L. Tora for support during the initial part of this work, S. Bour for graphic work, Y. Miyanari for advice on RNA FISH, E. Heard (Curie Institute, Paris, France) for the full-length LINE-1 probe, M. Dewannieux and T. Heidmann (Gustave Roussy Institute, Villejuif, France) for the IAP probe, D. Stollar, T. Ye and members of the Torres-Padilla lab for helpful discussions and A.J. Bannister and A. Burton for critical reading of the manuscript. M.-E.T.-P. acknowledges funding from AVENIR/INSERM, ANR-09-Blanc-0114, Epigenesys NoE and ERC-2011-StG 280840 “NuclearPotency.” Work in P.C.'s laboratory is supported by a Research Grant for RIKEN Omics Science Center from MEXT and a Funding Program for the Next Generation World Leading Researchers. A.F. was supported by a post-doctoral fellowship from the Fondation pour la Recherche Medicale.
Author information
Authors and Affiliations
Contributions
A.F. designed, performed and analyzed most of the experiments in this study; S.L.G. performed all bioinformatic analyses; B.J. and H.T. generated libraries; C.Z.-B. performed experiments; E.G. provided the triplex antibody; P.C. supervised library generation. M.-E.T.-P. conceived of the project, designed and supervised the study and performed experiments. A.F., S.L.G. and M.-E.T.-P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Tables 1 and 2 and Supplementary Note (PDF 3419 kb)
Rights and permissions
About this article
Cite this article
Fadloun, A., Le Gras, S., Jost, B. et al. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol 20, 332–338 (2013). https://doi.org/10.1038/nsmb.2495
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2495
This article is cited by
-
Fanconi anemia DNA crosslink repair factors protect against LINE-1 retrotransposition during mouse development
Nature Structural & Molecular Biology (2023)
-
N6-methyladenosine regulates maternal RNA maintenance in oocytes and timely RNA decay during mouse maternal-to-zygotic transition
Nature Cell Biology (2022)
-
Locus-specific expression of transposable elements in single cells with CELLO-seq
Nature Biotechnology (2022)
-
Retrotransposing a promoter for development
Nature Cell Biology (2021)
-
Stem cells-derived natural killer cells for cancer immunotherapy: current protocols, feasibility, and benefits of ex vivo generated natural killer cells in treatment of advanced solid tumors
Cancer Immunology, Immunotherapy (2021)