Abstract
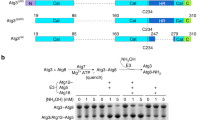
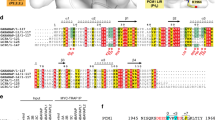
Autophagy requires ubiquitin-like Atg8 and Atg12 conjugation systems, where Atg7 has a critical role as the sole E1 enzyme. Although Atg7 recognizes two distinct E2s, Atg3 and Atg10, it is not understood how Atg7 correctly loads these E2s with their cognate ubiquitin-like proteins, Atg8 and Atg12. Here, we report the crystal structures of the N-terminal domain of Atg7 bound to Atg10 or Atg3 of thermotolerant yeast and plant homologs. The observed Atg7-Atg10 and Atg7-Atg3 interactions, which resemble each other but are quite distinct from the canonical E1-E2 interaction, makes Atg7 suitable for transferring Atg12 to Atg10 and Atg8 to Atg3 by a trans mechanism. Notably, in vitro experiments showed that Atg7 loads Atg3 and Atg10 with Atg8 and Atg12 in a nonspecific manner, which suggests that cognate conjugate formation in vivo is not an intrinsic quality of Atg7.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 (2011).
Nakatogawa, H., Suzuki, K., Kamada, Y. & Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 (2009).
Noda, N.N., Ohsumi, Y. & Inagaki, F. ATG systems from the protein structural point of view. Chem. Rev. 109, 1587–1598 (2009).
Schulman, B.A. & Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signaling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331 (2009).
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000).
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998).
Noda, N.N. et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell 44, 462–475 (2011).
Taherbhoy, A.M. et al. Atg8 transfer from Atg7 to Atg3: a distinctive E1–E2 architecture and mechanism in the autophagy pathway. Mol. Cell 44, 451–461 (2011).
Hong, S.B. et al. Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat. Struct. Mol. Biol. 18, 1323–1330 (2011).
Komatsu, M. et al. The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1–E2 complex formation. J. Biol. Chem. 276, 9846–9854 (2001).
Taherbhoy, A.M., Kaiser, S.E. & Schulman, B.A. Trans mechanism for ubiquitin-like protein transfer in autophagy. Cell Cycle 11, 635–636 (2012).
Yamada, Y. et al. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 282, 8036–8043 (2007).
Yamaguchi, M. et al. Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate. Structure 20, 1244–1254 (2012).
Huang, D.T. et al. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1. Mol. Cell 17, 341–350 (2005).
Yamaguchi, M. et al. Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 285, 29599–29607 (2010).
Fujioka, Y. et al. In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J. Biol. Chem. 283, 1921–1928 (2008).
Radoshevich, L. et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 142, 590–600 (2010).
Noda, N.N., Ohsumi, Y. & Inagaki, F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584, 1379–1385 (2010).
Xie, Z., Nair, U. & Klionsky, D.J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19, 3290–3298 (2008).
Nakatogawa, H., Ichimura, Y. & Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 (2007).
Hosokawa, N., Hara, Y. & Mizushima, N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 580, 2623–2629 (2006).
Hanada, T. et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302 (2007).
Kobashigawa, Y., Kumeta, H., Ogura, K. & Inagaki, F. Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method. J. Biomol. NMR 43, 145–150 (2009).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Brünger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Holm, L., Kaariainen, S., Rosenstrom, P. & Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008).
Delano, W. The Pymol Molecular Graphics System (Delano Scientific, 2002).
Kirisako, T. et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446 (1999).
Yoshimoto, K. et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983 (2004).
Suzuki, N.N., Yoshimoto, K., Fujioka, Y., Ohsumi, Y. & Inagaki, F. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy 1, 119–126 (2005).
Abramoff, M.D., Magalhaes, P.J. & Ram, S.J. Image Processing with ImageJ. Biophotonics International 11, 36–42 (2004).
Acknowledgements
The synchrotron radiation experiments were performed at beamlines BL41XU of SPring8 and BL5A, NE3A and NW12A of KEK in Japan. This work was supported in part by JSPS KAKENHI grant numbers 23687012 (to N.N.N.), 23000015 (to Y.O.), MEXT KAKENHI 24113725 (to N.N.N.) and MEXT Targeted Proteins Research Program (to F.I. and Y.O.).
Author information
Authors and Affiliations
Contributions
M.Y., K.M. and N.N.N. performed structural studies; M.Y., K.M., R.S., Y.F. and Y.K. performed biochemical studies; H.N. and H.Y. performed yeast experiments; H.H. and R.A. cloned Atg homologs from K. marxianus. M.Y., K.M., Y.O., N.N.N. and F.I. analyzed data and wrote the manuscript. All authors discussed the results and commented on the manuscript. N.N.N. and F.I. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 873 kb)
Rights and permissions
About this article
Cite this article
Yamaguchi, M., Matoba, K., Sawada, R. et al. Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7. Nat Struct Mol Biol 19, 1250–1256 (2012). https://doi.org/10.1038/nsmb.2451
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2451
This article is cited by
-
Multifaceted membrane interactions of human Atg3 promote LC3-phosphatidylethanolamine conjugation during autophagy
Nature Communications (2023)
-
An N-terminal conserved region in human Atg3 couples membrane curvature sensitivity to conjugase activity during autophagy
Nature Communications (2021)
-
NMR resonance assignments of human Atg3 in aqueous solution and bicelles
Biomolecular NMR Assignments (2021)
-
A switch element in the autophagy E2 Atg3 mediates allosteric regulation across the lipidation cascade
Nature Communications (2019)
-
Autophagy exacerbates electrical remodeling in atrial fibrillation by ubiquitin-dependent degradation of L-type calcium channel
Cell Death & Disease (2018)