Abstract
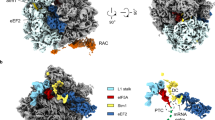
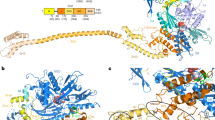
Ribosome-associated chaperones act in early folding events during protein synthesis. Structural information is available for prokaryotic chaperones (such as trigger factor), but structural understanding of these processes in eukaryotes lags far behind. Here we present structural analyses of the eukaryotic ribosome-associated complex (RAC) from Saccharomyces cerevisiae and Chaetomium thermophilum, consisting of heat-shock protein 70 (Hsp70) Ssz1 and the Hsp40 Zuo1. RAC is an elongated complex that crouches over the ribosomal tunnel exit and seems to be stabilized in a distinct conformation by expansion segment ES27. A unique α-helical domain in Zuo1 mediates ribosome interaction of RAC near the ribosomal proteins L22e and L31e and ribosomal RNA helix H59. The crystal structure of the Ssz1 ATPase domain bound to ATP-Mg2+ explains its catalytic inactivity and suggests that Ssz1 may act before the RAC-associated chaperone Ssb. Our study offers insights into the interplay between RAC, the ER membrane–integrated Hsp40-type protein ERj1 and the signal-recognition particle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Preissler, S. & Deuerling, E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 37, 274–283 (2012).
Kramer, G., Boehringer, D., Ban, N. & Bukau, B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 (2009).
Lowther, W.T. & Matthews, B.W. Structure and function of the methionine aminopeptidases. Biochim. Biophys. Acta 1477, 157–167 (2000).
Polevoda, B. & Sherman, F. Composition and function of the eukaryotic N-terminal acetyltransferase subunits. Biochem. Biophys. Res. Commun. 308, 1–11 (2003).
Grudnik, P., Bange, G. & Sinning, I. Protein targeting by the signal recognition particle. Biol. Chem. 390, 775–782 (2009).
Cross, B.C., Sinning, I., Luirink, J. & High, S. Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell Biol. 10, 255–264 (2009).
Gautschi, M. et al. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA 98, 3762–3767 (2001).
Hartl, F.U. & Hayer-Hartl, M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16, 574–581 (2009).
Bukau, B., Weissman, J. & Horwich, A. Molecular chaperones and protein quality control. Cell 125, 443–451 (2006).
Kampinga, H.H. & Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 (2010).
Gautschi, M., Mun, A., Ross, S. & Rospert, S. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA 99, 4209–4214 (2002).
Otto, H. et al. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc. Natl. Acad. Sci. USA 102, 10064–10069 (2005).
Huang, P., Gautschi, M., Walter, W., Rospert, S. & Craig, E.A. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol. 12, 497–504 (2005).
Yan, W. et al. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 17, 4809–4817 (1998).
Koplin, A. et al. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide–associated complex on ribosomes. J. Cell Biol. 189, 57–68 (2010).
Albanèse, V., Reissmann, S. & Frydman, J. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J. Cell Biol. 189, 69–81 (2010).
Karbstein, K. Chaperoning ribosome assembly. J. Cell Biol. 189, 11–12 (2010).
Amlacher, S. et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146, 277–289 (2011).
Fiaux, J. et al. Structural analysis of the ribosome-associated complex (RAC) reveals an unusual Hsp70/Hsp40 interaction. J. Biol. Chem. 285, 3227–3234 (2010).
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Mourão, A., Varrot, A., Mackereth, C.D., Cusack, S. & Sattler, M. Structure and RNA recognition by the snRNA and snoRNA transport factor PHAX. RNA 16, 1205–1216 (2010).
Peisker, K. et al. Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol. Biol. Cell 19, 5279–5288 (2008).
O'Brien, M.C., Flaherty, K.M. & McKay, D.B. Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis. J. Biol. Chem. 271, 15874–15878 (1996).
Sousa, M.C. & McKay, D.B. The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change. Biochemistry 37, 15392–15399 (1998).
Johnson, E.R. & McKay, D.B. Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein. Biochemistry 38, 10823–10830 (1999).
Beckmann, R. et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372 (2001).
Nilsson, J., Sengupta, J., Gursky, R., Nissen, P. & Frank, J. Comparison of fungal 80S ribosomes by cryo-EM reveals diversity in structure and conformation of rRNA expansion segments. J. Mol. Biol. 369, 429–438 (2007).
Armache, J.P. et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. Proc. Natl. Acad. Sci. USA 107, 19748–19753 (2010).
Melnikov, S. et al. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19, 560–567 (2012).
Blau, M. et al. ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nat. Struct. Mol. Biol. 12, 1015–1016 (2005).
Preissler, S. & Deuerling, E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 37, 274–283 (2012).
Schlenker, O., Hendricks, A., Sinning, I. & Wild, K. The structure of the mammalian signal recognition particle (SRP) receptor as prototype for the interaction of small GTPases with Longin domains. J. Biol. Chem. 281, 8898–8906 (2006).
Rakwalska, M. & Rospert, S. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol. Cell Biol. 24, 9186–9197 (2004).
Rospert, S., Rakwalska, M. & Dubaquie, Y. Polypeptide chain termination and stop codon readthrough on eukaryotic ribosomes. Rev. Physiol. Biochem. Pharmacol. 155, 1–30 (2005).
Halic, M. et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427, 808–814 (2004).
Amlacher, S. et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146, 277–289 (2011).
Collaborative Computational Project, No. 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Jia-Xing, Y., Woolfson, M.M., Wilson, K.S. & Dodson, E.J. A modified ACORN to solve protein structures at resolutions of 1.7 Å or better. Acta Crystallogr. D Biol. Crystallogr. 61, 1465–1475 (2005).
Langer, G., Cohen, S.X., Lamzin, V.S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Girish, V. & Vijayalakshmi, A. Affordable image analysis using NIH Image/ImageJ. Indian J. Cancer 41, 47 (2004).
Wagenknecht, T., Grassucci, R. & Frank, J. Electron microscopy and computer image averaging of ice-embedded large ribosomal subunits from Escherichia coli. J. Mol. Biol. 199, 137–147 (1988).
Shaikh, T.R. et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3, 1941–1974 (2008).
Norousi, R. et al. Automatic post-picking improves particle image detection from cryo-EM micrographs. Preprint at http://arxiv.org/abs/1112.3173v2 (2011).
Penczek, P.A., Frank, J. & Spahn, C.M. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G–dependent translocation. J. Struct. Biol. 154, 184–194 (2006).
Becker, T. et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373 (2009).
Putnam, C.D., Hammel, M., Hura, G.L. & Tainer, J.A. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 40, 191–285 (2007).
Konarev, P.V., Volkov, V.V., Sokolova, A.V., Koch, M.H.J. & Svergun, D.I. PRIMUS: a Windows PC–based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 (2003).
Svergun, D.I., Petoukhov, M.V. & Koch, M.H. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 (2001).
Volkov, V.V. & Svergun, D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 (2003).
Wriggers, W. Using Situs for the integration of multi-resolution structures. Biophys. Rev. 2, 21–27 (2010).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank A. Hendricks (BZH) for excellent technical assistance, C. Siegmann from the BZH Cluster of Excellence CellNetworks crystallization platform for technical support, and M. Rakwalska and R. Zimmermann for stimulating discussion. We thank O. Berninghausen and C. Ungewickell from the Ludwig Maximilians University Munich EM facility for their support, A. Bracher (Max Planck Institute for Biochemistry) for the Zuo1-N plasmid and S. Rospert for help with the ScRAC purification. X-ray data collection was performed at the European Synchrotron Radiation Facility (ESRF), and SAXS data were collected on the X33 beamline at the Deutsches Elektronen Synchotron–European Molecular Biology Laboratory (DESY/EMBL). We thank the staffs of both synchrotrons for excellent support. E.H. and I.S. are investigators of the Cluster of Excellence CellNetworks. G.B. is supported by the Peter and Traudl Engelhorn Foundation. R.B. and G.W. are funded by German Research Foundation (DFG) grant GRK1721. This work was supported by DFG grants to R.B. (FOR967 and SFB594) and I.S. (FOR967 and SFB638).
Author information
Authors and Affiliations
Contributions
C.L., G.B., R.B. and I.S. designed the experiments, analyzed the data and wrote the manuscript. G.B., A.A. and J.K. performed crystallographic analysis. C.L. and S.W. performed cryo-EM analysis. C.L. and G.W. performed SAXS analysis. S.A. and E.H. provided Ct80S ribosomes and cDNA of C. thermophilum. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 1589 kb)
Rights and permissions
About this article
Cite this article
Leidig, C., Bange, G., Kopp, J. et al. Structural characterization of a eukaryotic chaperone—the ribosome-associated complex. Nat Struct Mol Biol 20, 23–28 (2013). https://doi.org/10.1038/nsmb.2447
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2447
This article is cited by
-
Identification and characterization of sugar-regulated promoters in Chaetomium thermophilum
BMC Biotechnology (2023)
-
Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1
Nature Structural & Molecular Biology (2022)
-
Pathway of Hsp70 interactions at the ribosome
Nature Communications (2021)
-
The ribosome-associated complex RAC serves in a relay that directs nascent chains to Ssb
Nature Communications (2020)
-
HSP70L1-mediated intracellular priming of dendritic cell vaccination induces more potent CTL response against cancer
Cellular & Molecular Immunology (2018)