Abstract
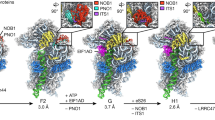
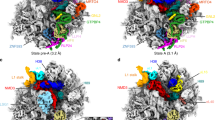
Eukaryotic ribosome biogenesis requires many protein factors that facilitate the assembly, nuclear export and final maturation of 40S and 60S particles. We have biochemically characterized ribosomal complexes of the yeast 60S-biogenesis factor Arx1 and late-maturation factors Rei1 and Jjj1 and determined their cryo-EM structures. Arx1 was visualized bound to the 60S subunit together with Rei1, at 8.1-Å resolution, to reveal the molecular details of Arx1 binding whereby Arx1 arrests the eukaryotic-specific rRNA expansion segment 27 near the polypeptide tunnel exit. Rei1 and Jjj1, which have been implicated in Arx1 recycling, bind in the vicinity of Arx1 and form a network of interactions. We suggest that, in addition to the role of Arx1 during pre-60S nuclear export, the binding of Arx1 conformationally locks the pre-60S subunit and inhibits the premature association of nascent chain–processing factors to the polypeptide tunnel exit.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Steitz, T.A. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 9, 242–253 (2008).
Rabl, J., Leibundgut, M., Ataide, S.F., Haag, A. & Ban, N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331, 730–736 (2011).
Klinge, S., Voigts-Hoffmann, F., Leibundgut, M., Arpagaus, S. & Ban, N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334, 941–948 (2011).
Ben-Shem, A. et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529 (2011).
Kressler, D., Hurt, E. & Bassler, J. Driving ribosome assembly. Biochim. Biophys. Acta 1803, 673–683 (2010).
Panse, V.G. & Johnson, A.W. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 35, 260–266 (2010).
Lempiäinen, H. & Shore, D. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 21, 855–863 (2009).
Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 (1999).
Strunk, B.S. et al. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333, 1449–1453 (2011).
Sengupta, J. et al. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J. Cell Biol. 189, 1079–1086 (2010).
Kramer, G., Boehringer, D., Ban, N. & Bukau, B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 (2009).
Gartmann, M. et al. Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J. Biol. Chem. 285, 14848–14851 (2010).
Hung, N.-J. & Johnson, A.W. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 26, 3718–3727 (2006).
Meyer, A.E., Hung, N.-J., Yang, P., Johnson, A.W. & Craig, E.A. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. USA 104, 1558–1563 (2007).
Giglione, C., Boularot, A. & Meinnel, T. Protein N-terminal methionine excision. Cell. Mol. Life Sci. 61, 1455–1474 (2004).
Hung, N.-J., Lo, K.-Y., Patel, S.S., Helmke, K. & Johnson, A.W. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol. Biol. Cell 19, 735–744 (2008).
Bradatsch, B. et al. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell 27, 767–779 (2007).
Kowalinski, E. et al. The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions. FEBS Lett. 581, 4450–4454 (2007).
Lebreton, A. et al. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 173, 349–360 (2006).
Demoinet, E., Jacquier, A., Lutfalla, G. & Fromont-Racine, M. The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae. RNA 13, 1570–1581 (2007).
Meyer, A.E., Hoover, L.A. & Craig, E.A. The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the pre-60 S subunit factor Arx1. J. Biol. Chem. 285, 961–968 (2010).
Lo, K.-Y. et al. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell 39, 196–208 (2010).
Merl, J. et al. Analysis of ribosome biogenesis factor-modules in yeast cells depleted from pre-ribosomes. Nucleic Acids Res. 38, 3068–3080 (2010).
Ho, J.H., Kallstrom, G. & Johnson, A.W. Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA 6, 1625–1634 (2000).
Nissan, T.A., Bassler, J., Petfalski, E., Tollervey, D. & Hurt, E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21, 5539–5547 (2002).
Pertschy, B. et al. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 27, 6581–6592 (2007).
Pettersen, E.F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Roderick, S.L. & Matthews, B.W. Structure of the cobalt-dependent methionine aminopeptidase from Escherichia coli: a new type of proteolytic enzyme. Biochemistry 32, 3907–3912 (1993).
Albanèse, V., Reissmann, S. & Frydman, J. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J. Cell Biol. 189, 69–81 (2010).
Sahi, C. & Craig, E.A. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. USA 104, 7163–7168 (2007).
Gautschi, M., Mun, A., Ross, S. & Rospert, S. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA 99, 4209–4214 (2002).
Yan, W. et al. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 17, 4809–4817 (1998).
Peisker, K. et al. Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol. Biol. Cell 19, 5279–5288 (2008).
Becker, T. et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373 (2009).
Halic, M. et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427, 808–814 (2004).
Beckmann, R. et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372 (2001).
Blau, M. et al. ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nat. Struct. Mol. Biol. 12, 1015–1016 (2005).
Bussiere, C., Hashem, Y., Arora, S., Frank, J. & Johnson, A.W. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J. Cell Biol. 197, 747–759 (2012).
Strunk, B.S., Novak, M.N., Young, C.L. & Karbstein, K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150, 111–121 (2012).
Lebaron, S. et al. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 19, 744–753 (2012).
Denning, D.P., Patel, S.S., Uversky, V., Fink, A.L. & Rexach, M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA 100, 2450–2455 (2003).
Jeeninga, R.E. et al. Variable regions V13 and V3 of Saccharomyces cerevisiae contain structural features essential for normal biogenesis and stability of 5.8S and 25S rRNA. RNA 3, 476–488 (1997).
Sweeney, R., Chen, L. & Yao, M.C. An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol. Cell. Biol. 14, 4203–4215 (1994).
Babiano, R. & de la Cruz, J. Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 38, 5177–5192 (2010).
Babiano, R., Gamalinda, M., Woolford, J.L. & de la Cruz, J. Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function. Mol. Cell. Biol. 32, 3228–3241 (2012).
van Beekvelt, C.A. et al. All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res. 29, 5001–5008 (2001).
Pöll, G. et al. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS ONE 4, e8249 (2009).
Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Schüler, M. et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 13, 1092–1096 (2006).
van Heel, M., Harauz, G., Orlova, E.V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Gabashvili, I.S. et al. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell 100, 537–549 (2000).
Valle, M. et al. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 21, 3557–3567 (2002).
Loerke, J., Giesebrecht, J. & Spahn, C.M.T. Multiparticle cryo-EM of ribosomes. Methods Enzymol. 483, 161–177 (2010).
Kelley, L.A. & Sternberg, M.J.E. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 (2009).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Jucker, F.M. & Pardi, A. Solution structure of the CUUG hairpin loop: a novel RNA tetraloop motif. Biochemistry 34, 14416–14427 (1995).
Schröder, G.F., Brunger, A.T. & Levitt, M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure 15, 1630–1641 (2007).
Honig, B. & Nicholls, A. Classical electrostatics in biology and chemistry. Science 268, 1144–1149 (1995).
Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. (2010).
Acknowledgements
We thank M. Leibundgut for critically reading this manuscript and E. Deuerling and T. Maier for discussions. We thank Electron Microscopy ETH Zurich (EMEZ) for data collection and P. Tittmann for support. This work was supported by the Swiss National Science Foundation (SNSF), the National Center of Excellence in Research (NCCR) Structural Biology program of the SNSF, and European Research Council (ERC) grant 250071 under the European Community's Seventh Framework Program (to N.B.).
Author information
Authors and Affiliations
Contributions
N.B., D.B. and B.J.G. conceived of the study. B.J.G. and C.M. performed biochemistry and sample preparation on the 60S-biogenesis factors and their complexes. B.J.G., D.B. and C.M. acquired cryo-EM data. B.J.G. calculated the cryo-EM structures with the help of D.B., and N.B., D.B. and B.J.G. interpreted the cryo-EM structures and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Note (PDF 699 kb)
Rights and permissions
About this article
Cite this article
Greber, B., Boehringer, D., Montellese, C. et al. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol 19, 1228–1233 (2012). https://doi.org/10.1038/nsmb.2425
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2425
This article is cited by
-
A molecular network of conserved factors keeps ribosomes dormant in the egg
Nature (2023)
-
Wide mutational analysis to ascertain the functional roles of eL33 in ribosome biogenesis and translation initiation
Current Genetics (2022)
-
Puf6 primes 60S pre-ribosome nuclear export at low temperature
Nature Communications (2021)
-
Arabidopsis REI-LIKE proteins activate ribosome biogenesis during cold acclimation
Scientific Reports (2021)
-
The roles of multifunctional protein ErbB3 binding protein 1 (EBP1) isoforms from development to disease
Experimental & Molecular Medicine (2020)