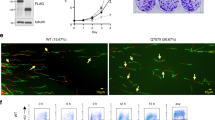
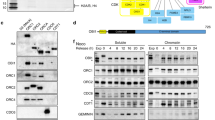
Abstract
Ubiquitin-binding domains (UBDs) are crucial for recruiting many proteins to sites of DNA damage. Here we characterize C1orf124 (Spartan; referred to as DVC1), which has an UBZ4-type UBD found predominantly in DNA repair proteins. DVC1 associates with DNA replication factories and localizes to sites of DNA damage in human cells, in a manner that requires the ability of the DVC1 UBZ domain to bind to ubiquitin polymers in vitro and a conserved PCNA-interacting motif. DVC1 interacts with the p97 protein 'segregase'. We show that DVC1 recruits p97 to sites of DNA damage, where we propose that p97 facilitates the extraction of the translesion synthesis (TLS) polymerase (Pol) η during DNA repair to prevent excessive TLS and limit the incidence of mutations induced by DNA damage. We introduce DVC1 as a regulator of cellular responses to DNA damage that prevents mutations when DNA damage occurs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Haglund, K. & Dikic, I. Ubiquitylation and cell signaling. EMBO J. 24, 3353–3359 (2005).
Ikeda, F. & Dikic, I. Atypical ubiquitin chains: new molecular signals. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 9, 536–542 (2008).
Hofmann, K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst.) 8, 544–556 (2009).
Sato, Y. et al. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 28, 2461–2468 (2009).
Sims, J.J. & Cohen, R.E. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol. Cell 33, 775–783 (2009).
Al-Hakim, A. et al. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst.) 9, 1229–1240 (2010).
Lehmann, A.R. Ubiquitin-family modifications in the replication of DNA damage. FEBS Lett. 585, 2772–2779 (2011).
Hoege, C., Pfander, B., Moldovan, G.L., Pyrowolakis, G. & Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 (2002).
Parker, J.L. & Ulrich, H.D. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J. 28, 3657–3666 (2009).
Kannouche, P.L., Wing, J. & Lehmann, A.R. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, 491–500 (2004).
Warbrick, E. PCNA binding through a conserved motif. BioEssays 20, 195–199 (1998).
Haracska, L. et al. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol. Cell. Biol. 21, 7199–7206 (2001).
Haracska, L. et al. Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc. Natl. Acad. Sci. USA 98, 14256–14261 (2001).
Haracska, L. et al. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol. Cell. Biol. 22, 784–791 (2002).
Watanabe, K. et al. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23, 3886–3896 (2004).
Bienko, M. et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310, 1821–1824 (2005).
Plosky, B.S. et al. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 25, 2847–2855 (2006).
Yang, W. & Woodgate, R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. USA 104, 15591–15598 (2007).
MacKay, C. et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142, 65–76 (2010).
Matic, I. et al. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39, 641–652 (2010).
Liu, Q. et al. Novel human BTB/POZ domain-containing zinc finger protein ZBTB1 inhibits transcriptional activities of CRE. Mol. Cell. Biochem. 357, 405–414 (2011).
Hooper, N.M. Families of zinc metalloproteases. FEBS Lett. 354, 1–6 (1994).
Centore, R.C., Yazinski, S.A., Tse, A. & Zou, L. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol. Cell 46, 625–635 (2012).
Machida, Y., Kim, M.S. & Machida, Y.J. Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell Cycle 11, 3395–3402 (2012).
Ghosal, G., Leung, J.W., Nair, B.C., Fong, K.W. & Chen, J. PCNA-binding protein C1orf124 is a regulator of translesion synthesis. J. Biol. Chem. advance online publication 17 August 2012 (doi:10.1074/jbc.M112.400135).
Taniguchi, T. et al. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100, 2414–2420 (2002).
Kratz, K. et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to unterstrand crosslinking agents. Cell 142, 77–88 (2010).
Shiomi, N. et al. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 35, e9 (2007).
Meerang, M. et al. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat. Cell Biol. 13, 1376–1382 (2011).
Acs, K. et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 18, 1345–1350 (2011).
Meyer, H., Bug, M. & Bremer, S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 (2012).
Zhang, Y. et al. Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res. 28, 4717–4724 (2000).
Parris, C.N. & Seidman, M.M. A signature element distinguishes sibling and independent mutations in a shuttle vector plasmid. Gene 117, 1–5 (1992).
Yeung, H.O. et al. Insights into adaptor binding to the AAA protein p97. Biochem. Soc. Trans. 36, 62–67 (2008).
Bienko, M. et al. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol. Cell 37, 396–407 (2010).
Maher, V.M., Ouellette, L.M., Curren, R.D. & McCormick, J.J. Frequency of ultraviolet light–induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature 261, 593–595 (1976).
Mosbech, A. et al. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat. Struct. Mol. Biol. advance online publication 7 October 2012 (doi:10.1038/nsmb.2395).
Acknowledgements
We thank A. Lehmann (University of Sussex) for Pol η expression plasmid, T. Shiomi (National Institute of Radiological Sciences, Japan) for HCT-116 RAD18−/− cells, M. Seidman (US National Institute on Aging, National Institutes of Health) for plasmid pSP189 and bacterial strain MBM7070, R. Toth (University of Dundee) for mutagenized DVC1 constructs, N. Mailand for communicating data before publication, and G. Alexandru and members of the Rouse laboratory for useful discussions. We thank the Medical Research Council Protein Phosphorylation Unit DNA Sequencing Service and the support teams of the Division of Signal Transduction Therapy, including N. Helps, J. Hastie and H. MacLauchlan, and members of the Centre for High Resolution Image Processing for help with microscopy and image processing. This study was funded by the Division of Signal Transduction Therapy Unit (funded by AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck KgaA, Janssen Pharmaceutica and Pfizer) associated with the Medical Research Council Protein Phosphorylation Unit, the Medical Research Council UK (E.J.D., C.L., T.J.M. and J.R.), the Association for International Cancer Research (E.J.D. and J.R.) and Cancer Research UK (P.A. and I.N.).
Author information
Authors and Affiliations
Contributions
J.R. designed the project, conceived the experiments and wrote the manuscript; E.J.D. and C.L. performed all of the experiments; T.J.M. made all of the plasmids used in this study; and P.A. and I.N. helped with microscopy.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Table 1 (PDF 1711 kb)
Rights and permissions
About this article
Cite this article
Davis, E., Lachaud, C., Appleton, P. et al. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat Struct Mol Biol 19, 1093–1100 (2012). https://doi.org/10.1038/nsmb.2394
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2394
This article is cited by
-
SPRTN patient variants cause global-genome DNA-protein crosslink repair defects
Nature Communications (2023)
-
DNA–protein crosslink proteases in genome stability
Communications Biology (2021)
-
DNA–protein cross-link repair: what do we know now?
Cell & Bioscience (2020)
-
TEX264 coordinates p97- and SPRTN-mediated resolution of topoisomerase 1-DNA adducts
Nature Communications (2020)
-
Polη O-GlcNAcylation governs genome integrity during translesion DNA synthesis
Nature Communications (2017)