Abstract
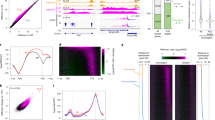
The plant-specific DNA-dependent RNA polymerase V (Pol V) evolved from Pol II to function in an RNA-directed DNA methylation pathway. Here, we have identified targets of Pol V in Arabidopsis thaliana on a genome-wide scale using ChIP-seq of NRPE1, the largest catalytic subunit of Pol V. We found that Pol V is enriched at promoters and evolutionarily recent transposons. This localization pattern is highly correlated with Pol V–dependent DNA methylation and small RNA accumulation. We also show that genome-wide chromatin association of Pol V is dependent on all members of a putative chromatin-remodeling complex termed DDR. Our study presents a genome-wide view of Pol V occupancy and sheds light on the mechanistic basis of Pol V localization. Furthermore, these findings suggest a role for Pol V and RNA-directed DNA methylation in genome surveillance and in responding to genome evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Law, J.A. & Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 (2010).
Cokus, S.J. et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219 (2008).
Haag, J.R. & Pikaard, C.S. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 12, 483–492 (2011).
Mosher, R.A., Schwach, F., Studholme, D. & Baulcombe, D.C. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. USA 105, 3145–3150 (2008).
Wierzbicki, A.T., Haag, J.R. & Pikaard, C.S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648 (2008).
Kanno, T. et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 14, 801–805 (2004).
Kanno, T. et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat. Genet. 40, 670–675 (2008).
Ausin, I., Mockler, T.C., Chory, J. & Jacobsen, S.E. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat. Struct. Mol. Biol. 16, 1325–1327 (2009).
Law, J.A. et al. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr. Biol. 20, 951–956 (2010).
Gao, Z. et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465, 106–109 (2010).
Zheng, B. et al. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 23, 2850–2860 (2009).
El-Shami, M. et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 21, 2539–2544 (2007).
Rowley, M.J., Avrutsky, M.I., Sifuentes, C.J., Pereira, L. & Wierzbicki, A.T. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet. 7, e1002120 (2011).
Huang, L. et al. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat. Struct. Mol. Biol. 16, 91–93 (2009).
Wierzbicki, A.T., Ream, T.S., Haag, J.R. & Pikaard, C.S. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41, 630–634 (2009).
Lee, T.F. et al. RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics 7, 798–795 (2012).
Ausin, I. et al. An IDN2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 109, 8374–8381 (2012).
Zhang, X., Bernatavichute, Y.V., Cokus, S., Pellegrini, M. & Jacobsen, S.E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10, R62 (2009).
Slotkin, R., Nuthikattu, S. & Jiang, N. The impact of transposable elements on gene and genome evolution, in Plant Genome Diversity Vol. 1 (ed. Wendel, J.F.) Ch. 3, 35–55 (Springer Wien, 2012).
Lisch, D. Epigenetic regulation of transposable elements in plants. Annu. Rev. Plant Biol. 60, 43–66 (2009).
Hollister, J.D. & Gaut, B.S. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19, 1419–1428 (2009).
Hu, T.T. et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476–481 (2011).
Hollister, J.D. et al. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc. Natl. Acad. Sci. USA 108, 2322–2327 (2011).
Pontier, D. et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19, 2030–2040 (2005).
Herr, A.J., Jensen, M.B., Dalmay, T. & Baulcombe, D.C. RNA polymerase IV directs silencing of endogenous DNA. Science 308, 118–120 (2005).
Johnson, L., Cao, X. & Jacobsen, S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12, 1360–1367 (2002).
Stroud, H. et al. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109, 5370–5375 (2012).
Lu, C., Meyers, B.C. & Green, P.J. Construction of small RNA cDNA libraries for deep sequencing. Methods 43, 110–117 (2007).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Chen, P.Y., Cokus, S.J. & Pellegrini, M.B.S. Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics 11, 203 (2010).
Spyrou, C., Stark, R., Lynch, A.G. & Tavare, S. BayesPeak: Bayesian analysis of ChIP-seq data. BMC Bioinformatics 10, 299 (2009).
Cairns, J. et al. BayesPeak—an R package for analysing ChIP-seq data. Bioinformatics 27, 713–714 (2011).
Acknowledgements
We thank T. Lagrange at the Université de Perpignan, Perpignan, France, for the NRPE1-Flag transgenic seeds and M. Akhavan for assistance with high-throughput sequencing. X.Z. is supported by Ruth L. Kirschstein National Research Service grant F32GM096483-01. C.J.H. is supported by the Damon Runyon Cancer Research Foundation fellowship. S.F. is supported by the Leukemia & Lymphoma Society Special fellowship. This work was supported by NIH grant GM60398, and S.E.J. is supported as an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
X.Z., C.J.H. and S.E.J. designed the experiments. X.Z., C.J.H., J.A.L., L.M.J., A.T. and S.F. performed the experiments. X.Z. and C.J.H. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 3–4 (PDF 1879 kb)
Supplementary Table 1
Genomic locations of identified NRPE1-enrichment sites and relative enrichment in NRPE1-FLAG ChIP-seq libraries compared to Col ChIP-seq libraries. (XLSX 405 kb)
Supplementary Table 2
“Ancient" transposons and corresponding blast hits between A. thaliana and A. lyrata. (XLSX 1072 kb)
Rights and permissions
About this article
Cite this article
Zhong, X., Hale, C., Law, J. et al. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol 19, 870–875 (2012). https://doi.org/10.1038/nsmb.2354
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2354
This article is cited by
-
MORC proteins regulate transcription factor binding by mediating chromatin compaction in active chromatin regions
Genome Biology (2023)
-
A cryo-EM structure of KTF1-bound polymerase V transcription elongation complex
Nature Communications (2023)
-
Histone chaperone ASF1 mediates H3.3-H4 deposition in Arabidopsis
Nature Communications (2022)
-
BPM1 regulates RdDM-mediated DNA methylation via a cullin 3 independent mechanism
Plant Cell Reports (2022)
-
Embryo CHH hypermethylation is mediated by RdDM and is autonomously directed in Brassica rapa
Genome Biology (2021)