Abstract
Enterovirus 71 (EV71) is a major agent of hand, foot and mouth disease in children that can cause severe central nervous system disease and death. No vaccine or antiviral therapy is available. High-resolution structural analysis of the mature virus and natural empty particles shows that the mature virus is structurally similar to other enteroviruses. In contrast, the empty particles are markedly expanded and resemble elusive enterovirus-uncoating intermediates not previously characterized in atomic detail. Hydrophobic pockets in the EV71 capsid are collapsed in this expanded particle, providing a detailed explanation of the mechanism for receptor-binding triggered virus uncoating. These structures provide a model for enterovirus uncoating in which the VP1 GH loop acts as an adaptor-sensor for cellular receptor attachment, converting heterologous inputs to a generic uncoating mechanism, highlighting new opportunities for therapeutic intervention.
Similar content being viewed by others
Main
Hand, foot and mouth disease (HFMD) is a serious public health threat across the Asia Pacific region, as evidenced by the >1.7 million cases in China reported by the Chinese Ministry of Health during 2010. The dominant causative agent is EV71, a nonenveloped single-stranded RNA virus (genus Enterovirus, family Picornaviridae) that is closely related to coxsackievirus A16, the other etiological agent of HFMD1. Whereas coxsackievirus A16 infections are not usually serious, acute EV71 infections can cause severe neurological disease2,3 and led to 905 deaths in China in 2010.
The icosahedral capsid of EV71 comprises 60 copies of four protein subunits, VP1–VP4. During assembly, the virally encoded P1 polyprotein is cleaved to yield VP0 (36 kDa), VP1 (32 kDa) and VP3 (27 kDa). VP0 is further processed into VP2 (28 kDa) and VP4 (8 kDa) in a reaction that is autocatalyzed by viral RNA and results in formation of the mature viral capsid4,5. In the mature capsid, VP1–VP3 follow a pseudo T = 3 arrangement and span the thickness of the capsid6; VP4 is located on the inside. Picornaviruses can also produce empty particles, which resemble the mature virus in structure and antigenicity, sediment at ~73S and comprise 60 copies of VP0, VP1 and VP3 (refs. 4,7,8). Inline with this, EV71 cultured for vaccine development produces two distinct types of particle9: highly infectious mature virions composed of VP1–VP4 and RNA, and empty particles containing VP0, VP1 and VP3. Although 73S particles are no longer thought to be direct precursors of mature virions, they can function as reservoirs of capsid components8,10,11, presumably following their dissociation into functional subunits. 73S particles are unstable, readily converting from D-type native antigenicity to a state with altered (C-type in the poliovirus nomenclature12,13) antigenic properties.
Structural studies have outlined the processes leading to enteroviral infection of cells14. Cellular receptors attach to the virus, often binding in a canyon-like depression surrounding the five-fold axis15,16,17. This triggers conformational changes in the virus, resulting in the formation of an expanded intermediate that sediments at ~135S (as compared to about ~160S for the mature virus) and has altered antigenic properties18,19,20. These conformational changes (which can also be induced by environmental insults such as heating or low pH) lead to the externalization of VP4 and the VP1 N terminus21,22. This is followed by extrusion of the viral genome into the cytoplasm of the target cell, leaving behind an empty ~80S particle21.
Previous low-resolution data indicated that the viral RNA probably exits through a five-fold axis channel14, although recent studies suggest that it exits near a two-fold axis23. Nevertheless, there remains a lack of high-resolution structural detail to illuminate the molecular mechanisms underpinning the dynamic enterovirus cell-entry process. To address this, we have determined structures of both inactivated and infectious EV71 virions. We have also determined structures of expanded natural empty particles that closely resemble the expanded enterovirus uncoating intermediates previously visualized by cryo-EM24,25. Our high-resolution analyses suggest a detailed molecular mechanism for the early stages of enterovirus uncoating.
Results
Characterization and structure determination
EV71 was isolated from a patient from Anhui, China, and cultivated in Vero cells. When required, the virus was inactivated using formaldehyde before purifying it by centrifugation, ultrafiltration, PEG precipitation and gel filtration (see Online Methods). Unless otherwise stated, analyses used inactivated virus. We separated out the two particle types of interest by ultracentrifugation and characterized them using SDS-PAGE and analytical ultracentrifugation as either 150S mature virions, containing RNA and a full complement of proteins VP1–VP4, or 82S empty particles containing VP0, VP1 and VP3 (Supplementary Fig. 1).
The sedimentation coefficient of the empty particles was considerably greater than that of native-antigenicity enterovirus empty particles (~73S), despite an earlier report that EV71 empty particles were smaller than the virus9. Furthermore, a monoclonal antibody recognized the virus but not the empty particle, indicating differences in antigenicity between the two particles (Supplementary Fig. 2). These results, and the considerable thermal stability of the empty particles (Supplementary Fig. 2), suggest that native-antigenicity EV71 empty particles rapidly and spontaneously reconfigure to the altered state that we characterize here.
We collected diffraction data from small crystals primarily at 21 °C directly from the crystallization plates (in situ), avoiding the problems of harvesting and cryoprotecting the fragile crystals. We believe this represents the first report of determination of a virus structure using such a method. We solved the structure of the mature inactivated virus in two space groups with data merged from multiple crystals. For the empty particle, we collected a high-resolution data set at 100 K and a lower-resolution data set at 21 °C. This provided reliable atomic models at resolutions of 2.3 Å, 2.6 Å, 2.9 Å and 3.8 Å, respectively, for two independent structure determinations each for full virus and empty particles (see Online Methods; Table 1 and Supplementary Fig. 3). Although particles had been inactivated with formaldehyde, there was no evidence in the electron density for specific covalent cross-linking occurring in a systematic manner, as inactivation did not seem to cross-link the majority of the capsid proteins either to each other or to viral RNA (Supplementary Fig. 1). Subsequent structure determinations of non-inactivated particles (Table 1) showed that inactivation did not perturb the structures and that inactivation presumably operates by the cooperative effect of a few cross-links made in each virus particle. Unless otherwise stated, descriptions below apply to all the structures of that particle type.
Structure of the mature virus
Mature EV71 is well ordered, apart from a few disordered residues in VP4 and VP2 (Fig. 1), and is similar to bovine enterovirus (BEV1), although the inner capsid surface is distinct from that of other enteroviruses (Supplementary Fig. 4). Notably, whereas in structurally characterized enteroviruses residues 1–28 of VP1 proceed toward the five-fold axis, in EV71 they veer across the protomer, presenting a short helix (residues 5–9) underneath the αA helices of VP2, adjacent to the icosahedral two-fold axes. Additionally, residues 14–31 of VP4 form a loose spiral beneath VP1 rather than lying under the adjacent biological protomer. One RNA molecule base-stacks against VP2 Trp38 (as in other enteroviruses26), and another may interact with VP1 Gln30 and VP3 Gln48. On the outside of the particle, the VP1 BC, DE and HI loops are flattened away from the five-fold axis (as in BEV1), with residues 96–102 (BC loop) and residues 208–222 (GH loop) being the most exposed (Fig. 1c). Part of the VP2 EF loop (residues 128–148) is unusually extended and surface dominant, whereas the remainder (residues 151–172) is shorter than usual and less accessible. Exposed residues in VP3 include 58–69 and 173–190 (Supplementary Fig. 5).
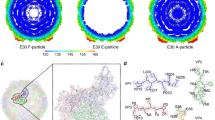
(a) Cartoon of the mature EV71 virion, looking down an icosahedral two-fold axis, VP1, VP2, VP3 and VP4 are drawn in blue, green, red and yellow, respectively. A single icosahedral protomer is drawn more brightly. (b) Radius-colored surface representation of the EV71 mature and empty particles. The surfaces for both are colored from blue to red according to their distances from the particle center (blue being closest). (c) Structures of the EV71 capsid proteins. VP0–VP3 (we treat VP0 and VP2 interchangeably) are shown in a similar orientation to the brighter protomer in a; proteins from the expanded empty particle are colored as in a, whereas the corresponding chains from the mature virion are gray. The proteins have been superimposed as rigid bodies. Residues 1–297 of VP1, 10–254 of VP2, 1–242 of VP3 and 12–69 of VP4 have well-defined electron density in the mature virion; residues 73–210 and 219–297 of VP1, 82–319 of VP0 and 1–238 of VP3 in the empty particle are modeled. Major surface-exposed loops are marked with colored circles: yellow, VP1 BC; orange, VP1 DE; red, VP1 HI; magenta, VP1 GH; light blue, VP2 EF; black, VP3 GH.
An expanded natural empty particle
The EV71 empty particles are markedly larger than any other picornavirus particle characterized crystallographically (Fig. 1b), with the r.m.s. capsid radius increasing from 132 Å for the mature virion to 139 Å for the empty particle. Whereas cooling virus particles to 100 K often induces modest isotopic shrinkage (~1%), both the 293-K and 100-K empty particles are ~4% larger than the 293-K mature virus. The 100-K particles are not icosahedral, so refinement imposing icosahedral symmetry stuck at an R factor of 30%, but it improved markedly when this constraint was relaxed. Cryo-cooling pushes Cαs in the crystal's contact region ~1.3 Å toward the particle center, but the 293-K and 100-K structures are otherwise essentially identical (Supplementary Fig. 5). Such deviations from icosahedral symmetry have never been observed in mature viruses and reflect the extreme flexibility of these expanded particles, explaining why it has been difficult to visualize them at high resolution.
The expansion of the empty particles reflects tectonic movements in the particle, partially separating the protomeric units, disordering >70,000 protein atoms per particle and forming perforations at the icosahedral two-fold axes and at the base of the canyon (Fig. 2a,b). Several external loops, which nestle at the junction of polypeptide chains, become disordered, including five residues at the C terminus of VP2 and residues 211–217 of the GH loop of VP1. The VP3 GH loop also undergoes a major conformational switch, with residues 170–192 converting from loop and helix to almost a β-hairpin upon expansion, becoming less ordered (for residues 174–190, B factors exceed 100 Å2) (Supplementary Fig. 5). Inside the particle, the internal festoon comprising VP1's N-terminal 72 residues and the first 81 residues of VP0 is now disordered (Fig. 1). Overall surface properties are appreciably altered, the interactions that hold the particle together are strongly reduced (Supplementary Table 1) and the capsid in the expanded particle is thinned to 20 Å from 23 Å in the mature virus (Fig. 2c). The core structures of the individual polypeptides are less affected as Cαs superpose with r.m.s. deviations of 1.6 Å, 0.9 Å and 1.3 Å for VP1, VP0(2) and VP3, respectively (Fig. 1c), although some important rearrangements occur.
(a,b) Averaged 2Fo – Fc maps in the vicinity of an icosahedral two-fold axis (marked as a black ellipse) for the mature virus (a) and the expanded particle (b), showing the well-defined electron density and the perforations in the expanded particle. Cα traces, colored as in Figure 1, are also shown. (c) Equivalent equatorial slices through the two electron-density maps shown a and b. Black lines mark the approximate position of an icosahedral five-fold axis. (d–g) Comparisons of the fit of our crystallographic EV71 expanded-particle coordinates and modeled poliovirus into the 10-Å resolution cryo-EM reconstructions for the 135S and 82S poliovirus particles24,25. The density was displayed and correlation coefficients calculated using the program URO28 for fitting of EV71 (d, green) and poliovirus (e, red) into the density for the 135S particle (correlation coefficient 0.66 for both EV71 and the deposited poliovirus coordinates; PDB 1XYR24). Also shown are the corresponding fits into the cryo-EM density for the late 80S poliovirus (correlation coefficients 0.63 for EV71 (f) and 0.64 for poliovirus (g)).
Enteroviruses adopt two fundamental configurations
The ~4% expansion of the EV71 82S empty particle is similar to that seen for the poliovirus 135S and 80S uncoating intermediates27, and the latter has similar hydrodynamic properties. Cryo-EM analyses revealed that the poliovirus 135S and 80S particles are structurally similar to each other27 (although the 80S particle has shed the viral genome) and are reconfigured compared to the mature virus24,25. We used VEDA28 to compare the EV71 expanded particle with the poliovirus particles. Unexpectedly, the EV71 particle fit the poliovirus 135S and 80S electron densities as well as the poliovirus structure previously modeled into this density (Fig. 2d–g); thus, at low resolution the EV71 expanded-assembly byproduct is indistinguishable from poliovirus uncoating intermediates. Furthermore, heat treatment of mature EV71 particles (using a protocol similar to that which produces 135S poliovirus particles) converts them to particles that crystallize isomorphously with 82S immature particles (data not shown). It therefore seems that the plethora of enterovirus particles formed during assembly and uncoating possess only two fundamental configurations, which are both now defined in atomic detail. The mature virus particle, which is rigid and rendered more stable by cleavage of VP0, is generally converted to the second, expanded configuration after cell attachment. This flexible expanded particle can adopt subtly different conformations during the process of uncoating as it progresses from 135S to 80S form25, but it can also arise from the conversion of unstable VP0 containing natural empty particles, as seen for poliovirus10,11 and now EV71.
Mechanics of particle expansion
Particle expansion is accompanied by a 5.4° counterclockwise rotation of the protomeric building block (VP1, VP0, VP3), which pivots about the corner of VP3 at the icosahedral three-fold axis (Fig. 3a). This screw-like movement moves VP1 and the underlying VP3 hub away from the particle center by up to 7 Å, and pulls VP0 away from the two-fold axis by 5 Å, opening up perforations. Residues 136–141 of VP3 and residues 227–250 and 48–52 of VP2 form a bridge separating the perforations.
(a) Protomeric units shown with respect to the icosahedral axes of the particles (that is, superposing whole particles). The mature virus is gray and the expanded particle is colored as in Figure 1c. The orientation is similar to that of the bright protomer in Figure 1a; icosahedral symmetry axes are drawn in black. (b) Superposed protomers51. Structural differences are mapped onto the protomer of the mature virion; the thickness and color of the worm representation reflect the local deviation between the structures (from blue (0 Å) to orange (8 Å)). Regions missing in the expanded particle are shown in red (VP4 is omitted). (c) The change in the jackknife structure of VP1 on capsid expansion. The superposition is based on VP0(2), VP3 and the five-fold distal portion of VP1. The black dot in the right diagram marks the point about which VP1 flexes. (d–g) Regions where perforations appear in the expanded particle (the base of the canyon in d and e and the two-fold region in f and g. d,f, mature virus; e,g, expanded particle. In c–g, colors are as in Figure 1c, with the GH loops of VP1 and VP3 highlighted in d and e in gray and magenta, respectively.
In addition to large-scale disordering (described above), small-scale changes occur on the edges of the protomers (Fig. 3b,c). The separation at the two-fold axes rips apart the αA helices of adjacent VP2 subunits that normally form a key interaction stabilizing the mature virus (Fig. 3d–g). In the expanded capsid, the C-terminal stabilizing helix cap (Arg249) becomes poorly ordered, suggesting mechanical strain (the five C-terminal residues beyond it are disordered). Within the protomeric unit, a 7.5° rotation and a 1.4-Å translation of the five-fold proximal end of the VP1 β-barrel, which is jackknifed upward in the mature virus (Fig. 3c), straightens the protomer and moves the GH loop, H strand, CD loop and residues ~262–280 beyond strand I of VP1. The trigger for protomer extension seems to be changes centered on a pocket in the VP1 β-barrel.
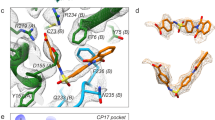
Mature EV71, as with other enteroviruses, possesses a hydrophobic pocket that penetrates from the surface deep into the interior of the VP1 β-barrel, underlying a canyon-like surface depression16 and harboring a natural lipid (possibly sphingosine, as seen in poliovirus29; Fig. 4 and Supplementary Fig. 3). Notably, the only visible difference between active and inactivated particles is that the occupancy of the 'pocket factor' in the active virus is approximately half that of the inactive virus (Supplementary Fig. 6). This suggests that formaldehyde treatment rigidifies the particle, inhibiting the release of the pocket factor and presumably contributing to inactivation, as pocket-factor release seems to be required for the initiation of uncoating30,31 (potential antivirals have been discovered that replace natural lipids and inhibit uncoating31,32). Until now, empty pockets observed in enteroviruses16,26,33,34 have been largely open (such that a pocket factor might bind without appreciable structural change). In contrast, not only is the EV71 expanded-particle pocket empty, but small conformational changes in its external walls have collapsed it from 1,000 Å3 to 580 Å3 (program Volumes; R. Esnouf, unpublished data) so that it can no longer accommodate lipids (Fig. 4b,c and Supplementary Figs. 4 and 6).
(a) The hydrophobic pocket (blue mesh) in VP1 of mature EV71 is occupied by a natural lipid (magenta). The protein cartoon is colored as in Figure 1a. (b) The expanded particle, showing the empty, collapsed pocket. (c) Comparison of the VP1 pockets between the mature virus (gray, with pocket factor shown in magenta) and empty particles (blue).
During pocket collapse, several residues move inward, taking some internal hydrophobic residues with them. The largest movement of the polypeptide backbone is in residues 230–233 at the end of the GH loop and the start of strand H, where Cα movements reach ~2.5 Å, and the Met230 and Phe233 side chains reposition to ablate the pocket (Fig. 4c). Notably, this portion of chain, which we term the 'adaptor-sensor', is directly downstream of a region of the GH loop that is external and structurally variable in poliovirus (Supplementary Fig. 6). This variability may reflect an unknown biological function or may simply facilitate the expansion switch. Rearrangement of the adaptor-sensor shifts the walls of the canyon to straighten the VP1 core. Furthermore, in mature particles this region directly contacts the C-terminal residues of VP2, including the Arg249 cap stabilizing the VP2 αA helix. In the expanded particle, this region becomes distorted and partly disordered, perhaps indicating strain that facilitates the separation of the VP2 αA helices to complete particle expansion. Indeed, a capping arginine is conserved in all enteroviruses (and cardioviruses) but absent from the less stable aphthoviruses (Supplementary Fig. 4). In summary, closure of the VP1 pocket initiates enterovirus uncoating via a mechanical connection to the pentamer interface.
Antigenicity and receptor engagement
Residues 210–220 of VP1, preceding the adaptor-sensor, are part of an important neutralizing epitope of EV71 (ref. 35). They lie on the capsid surface, alongside the VP2 EF loop (residues 136–150), to form a single epitope35,36; however, residues 211–217 become disordered upon particle expansion, probably explaining the loss of immunogenicity reported previously9 and seen by us (Supplementary Fig. 2). The surface charge of the virion is consistent with initial cell attachment occurring via a sugar moiety (Supplementary Fig. 7); three glycoproteins have been suggested as receptors37,38,39, but their binding sites on EV71 are unknown. Nevertheless, enterovirus receptors frequently attach at the canyon40, and mapping cryo-EM structures of other enterovirus-receptor complexes (PDB 1NN8 (ref. 41), PDB 1Z7Z42, PDB 1AYN43 and PDB 1MEC44) onto mature EV71 shows that they make similar contacts with the canyon walls at the adaptor-sensor region. We propose that full engagement of the receptor with the adaptor-sensor switches the adaptor-sensor's conformation to a high-receptor-affinity form (reported, for example, for poliovirus45), triggering expulsion of the pocket factor and conversion to the expanded particle.
Discussion
The mechanism by which the RNA genome is productively released from picornaviruses has long been the subject of speculation16,23,25,46, and, although the details will vary, the process is likely to be fundamentally similar for all enteroviruses. RNA release in poliovirus is preceded by the egress of the VP1 N terminus and VP4 (ref. 21), which may associate to form a channel through the membrane, allowing the safe transfer of the viral genome to the cell cytoplasm. Candidates for membrane association and channel formation are the myristate group at the N terminus of VP4 (ref. 47) and a region at the N terminus of VP1 that is proposed to form an amphipathic helix48. It had been assumed that a five-fold channel would be formed to release these regions46, but recent studies suggest instead that VP1 (and possibly VP4) leaves at the base of the canyon or at a two-fold axis23,24,49. We have defined a channel at the base of the canyon (7 × 9 Å in size) and a larger (8 × 25 Å) opening at the two-fold axis (Fig. 2b), either of which would allow egress of a polypeptide chain while the larger might also allow the exit of RNA.
In the mature EV71 virus, the N-terminal region of VP1 lies at the two-fold axis, rather than close to the five-fold axis, perhaps presenting a snapshot of a structural rearrangement that precedes VP1 release in other enteroviruses. Indeed, in the expanded particle, both VP4 and the VP1 N terminus are disordered, and possibly already partly extruded. During exit of the RNA, less structured regions of the single-stranded RNA genome might be transiently unwound through interactions with the inner surface of the capsid. However, the considerable secondary structure at the termini of picornavirus genomes suggests that strain would be required to initiate RNA egress, perhaps accounting for the fact that the RNA does not spontaneously exit 135S particles. This strain could probably be accommodated by the flexibility of the expanded particle, which could allow the two openings to merge by breaking the single weak link between them (Fig. 3e,g). Notably, flexing of this region would render a conserved VP3 sequence (PPGxxxPxxR50, reminiscent of a protein-binding motif) accessible on the inside of the particle. This motif may be involved in picornavirus assembly or uncoating.
Our results provide two lessons for vaccine production: (i) formaldehyde inactivation leaves the structure, and hence antigenicity, of EV71 essentially unchanged; (ii) when grown in Vero cells, this strain of EV71 produces large amounts of antigenically altered particles, which will, at best, dilute the useful portion of a vaccine. More broadly, our results, with those of others, show that the key structural transitions of enteroviruses are based on just two fundamental states, with different physicochemical and antigenic properties that are matched to their distinct roles in the virus life cycle. Atomic-level descriptions of these states allow us to propose a mechanism by which receptor binding triggers virus uncoating and to highlight points where we might attempt to modulate the switch between the two states, using either small molecules or mutant viruses. One such point is the well-studied VP1 hydrophobic pocket31. A second is the pair of VP2 αA helices, which are torn apart during expansion; mutations that weaken the link from the adaptor-sensor to this point might be useful in providing stabilized vaccines. Furthermore, key amino acid residues in the proposed uncoating mechanism are conserved beyond the enteroviruses (Supplementary Fig. 4), suggesting that, by targeting elements of the mechanism that are common to other picornavirus genera, it may be possible to develop generic therapies against a number of viruses posing serious threats to health.
Methods
Particle production and purification.
EV71 (genotype C4), isolated from Fuyang, Anhui Province, China, was used to infect Vero cells at 107 50% tissue culture infective dose (TCID50). The virus was collected 5–6 d after infection, inactivated by incubation with 100 μg ml−1 formaldehyde at 37 °C for 3 d, centrifuged to remove cell debris, ultrafiltered and PEG precipitated and subjected to gel filtration. These stages were performed by Sinovac Biotech Ltd and the China National Biotech Group. Crude EV71 concentrate (~0.6 mg in 600 μl PBS (pH 7.4)) was loaded onto a 15–45% (w/v) sucrose density gradient and centrifuged at 103,614g for 3.5 h in an SW41 rotor at 4 °C. Two sets of fractions were collected and dialyzed against PBS buffer (Supplementary Fig. 1); one comprised empty particles (containing no RNA), the other contained virions. SDS-PAGE analysis used a NuPAGE 4–12% Bis-Tris Gel (Invitrogen) (Supplementary Fig. 1).
Analytical ultracentrifugation.
The sedimentation coefficients for both types of particles were determined using a Beckman XL-I analytical ultracentrifuge at 4 °C (Supplementary Fig. 1).
Antibody D6 antigen-binding (Fab) purification and EV71 immunogenicity.
Anti-EV71 monoclonal antibody (mAb) D6 was supplied by Sinovac. Fab fragments were generated (Pierce@FAB preparation kit, Thermo Scientific), dialyzed against 20 mM acetate (pH 5.0) at 4 °C, loaded onto a Mono S column (GE Healthcare) and eluted using a 0–500 mM NaCl gradient. The main peak was collected and dialyzed into PBS buffer. The pure Fab was incubated with the semipurified EV71 sample containing empty and full particles (at a ratio of ~240 Fab molecules per EV71 virion) at 4 °C for 12 h. The mixture was loaded onto a 15–45% (w/v) sucrose density gradient and centrifuged at 103,614g for 3.5 h using a SW41 rotor at 4 °C (Supplementary Fig. 2).
Thermofluor assay.
Thermofluor experiments were performed with an MX3005p RT-PCR instrument (Agilent). SYTO9 and SYPROred (both Invitrogen) were used as fluorescent probes to detect the presence of RNA and the exposed hydrophobic regions of proteins, respectively. We set up 50-μl reactions in a thin-walled PCR plate (Agilent), containing 0.5–1.0 μg of either the virus or empty particles, 5 μM SYTO9 and 3X SYPROred in PBS (pH 7.4) and ramped the temperature from 25 °C to 99 °C, with fluorescence recorded in triplicate at 1 °C intervals. The melting temperature, Tm, was taken as the minimum of the negative first derivative of the denaturation curve (Supplementary Fig. 2).
Crystallization.
Crystallization used nanoliter vapor diffusion in Greiner CrystalQuick X plates52. Purified particles were concentrated to 2mg ml−1 in PBS (pH 7.4). Crystal Screen1 (Hampton Research) condition 13 (30% (v/v) PEG400, 0.2 M tri–sodium citrate, 0.1 M Tris-HCl (pH 8.5)) gave small irregular crystals. Because standard optimization techniques failed, 20% of this condition was added to the virus solution and then equilibrated against alternative reservoirs: the SaltRx screen solutions (Hampton Research). Crystals emerged in more than half of the 96 conditions with maximum size of 0.15 × 0.15 × 0.02 mm3, in three crystal morphologies: rhomboid plates, and cubic and triangular prisms. Crystals of empty particles grew in two conditions: (i) 200 mM ammonium phosphate monobasic, 24% (v/v) isopropanol and 100 mM sodium cacodylate (pH 6.5); (ii) 1.4 M sodium acetate and 0.1 M sodium cacodylate (pH 6.5). The crystallization conditions were similar for inactivated and non-inactivated virus.
Structure determination.
Diffraction data were taken at room temperature (293 K) from crystals in crystallization plates (in situ), a method developed at beamline I24, Diamond53. Diffraction images of 0.05° or 0.1° oscillation were recorded on a Pilatus 6M detector using a 0.02 × 0.02 mm2 or 0.05 × 0.05 mm2 beam according to crystal size. The X-ray beam was focused downstream from the crystal. Using a 0.05-s exposure time and 100% beam transmission, typically six to ten useful images could be collected from one position on a crystal (up to six positions for larger crystals). Data for the EV71 inactivated full particles were collected from the rhomboid plates and cubic crystals (space groups R32 and I23, respectively; Table 1). Room temperature data were also collected for the empty EV71 inactivated particles to 3.8-Å resolution. The data were weak (by the usual current standards of macromolecular crystallography), and this is reflected in the poor merging R factors. Subsequently, data were collected, to lower resolution, from non-inactivated full and empty particles (Table 1).
Cryo-cooled (100 K) data for crystals of empty EV71 particles were measured using 0.1° 1-s exposure oscillations. Crystals were soaked for 20 min in a cryoprotectant solution (80% (v/v) reservoir solution and 20% (v/v) glycerol) before being plunged into liquid nitrogen.
Data were analyzed using HKL2000 (ref. 54). The structures of both empty and full inactivated particles were determined by molecular replacement. For the empty particle (P4232, a = b = c = 353.1 Å (100 K data)) there is an icosahedral pentamer in the crystallographic asymmetric unit. For the full virus (R32 a = b = 330.0 Å and c = 748.4 Å, and I23 a = b = c = 600.1 Å) there are two and four pentamers, respectively, in the asymmetric unit. Molecular replacement used BEV (PDB 1BEV)34 as the search molecule (most orientation and positional parameters were defined by the alignment of icosahedral and crystallographic symmetry axes). For the I23 crystals, the center of the virus is at (0.25, 0.25, 0.25). In each case, the crystallographic asymmetric unit was rigid-body refined. Cyclic positional, simulated annealing and B-factor refinement used strict NCS constraints with CNS55. Averaging was done using GAP (D.I.S., J. Grimes and J. Diprose, unpublished data) and models were rebuilt with COOT56. Refinement of the empty-particle cryo-structure stuck at an R factor of 0.30. Rigid-body refinement of individual protomers (breaking the icosahedral symmetry) reduced the R factor to 0.25 (all data to 2.88 Å). Inspection revealed that, at the crystallographic contacts, the structure was squashed 1.3 Å inward, presumably as a result of crystal freezing. BUSTER57 refinement using NCS restraints gave an R factor of 0.187. Recalculated NCS matrices were used as constraints with CNS, resulting in the final structure (Table 1). The non-inactivated structures were isomorphous with the inactivated structures and were refined directly from those structures, using strict NCS constraints in CNS. Models were verified with PROCHECK58 (Table 1). Structural comparisons used SHP51. Unless otherwise noted, structural figures were prepared with PyMOL (DeLano Scientific).
Accession codes.
Protein Data Bank: Coordinates and structure factors for formaldehyde-inactivated full virus in two space groups, the empty particle at 100 K and 21 °C, and native full and empty viruses have been deposited with accession codes 3VBF, 3VBH, 3VBO, 3VBR, 3VBS and 3VBU, respectively.
References
Brown, B.A. & Pallansch, M.A. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 39, 195–205 (1995).
McMinn, P.C. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 26, 91–107 (2002).
Wu, Y. et al. Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China. Protein Cell 1, 491–500 (2010).
Basavappa, R. et al. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 resolution. Protein Sci. 3, 1651–1669 (1994).
Curry, S. et al. Dissecting the roles of VP0 cleavage and RNA packaging in picornavirus capsid stabilization: the structure of empty capsids of foot-and-mouth disease virus. J. Virol. 71, 9743–9752 (1997).
Tuthill, T.J., Groppelli, E., Hogle, J.M. & Rowlands, D.J. Picornaviruses. Curr. Top. Microbiol. Immunol. 343, 43–89 (2010).
Ansardi, D.C. & Morrow, C.D. Amino acid substitutions in the poliovirus maturation cleavage site affect assembly and result in accumulation of provirions. J. Virol. 69, 1540–1547 (1995).
Jacobson, M.F. & Baltimore, D. Morphogenesis of poliovirus. I. Association of the viral RNA with coat protein. J. Mol. Biol. 33, 369–378 (1968).
Liu, C.C. et al. Purification and characterization of enterovirus 71 viral particles produced from Vero cells grown in a serum-free microcarrier bioreactor system. PLoS ONE 6, e20005 (2011).
Guttman, N. & Baltimore, D. A plasma membrane component able to bind and alter virions of poliovirus type 1: studies on cell-free alteration using a simplified assay. Virology 82, 25–36 (1977).
Marongiu, M.E., Pani, A., Corrias, M.V., Sau, M. & La Colla, P. Poliovirus morphogenesis. I. Identification of 80S dissociable particles and evidence for the artifactual production of procapsids. J. Virol. 39, 341–347 (1981).
Le Bouvier, G.L. The modification of poliovirus antigens by heat and ultraviolet light. Lancet 269, 1013–1016 (1955).
Blondel, B., Akacem, O., Crainic, R., Couillin, P. & Horodniceanu, F. Detection by monoclonal antibodies of an antigenic determinant critical for poliovirus neutralization present on VP1 and on heat-inactivated virions. Virology 126, 707–710 (1983).
Hogle, J.M. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 56, 677–702 (2002).
Olson, N.H. et al. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. USA 90, 507–511 (1993).
Rossmann, M.G. et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317, 145–153 (1985).
Zhang, P. et al. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc. Natl. Acad. Sci. USA 105, 18284–18289 (2008).
Crowell, R.L. & Philipson, L. Specific alterations of coxsackievirus B3 eluted from HeLa cells. J. Virol. 8, 509–515 (1971).
Kaplan, G. et al. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J. 15, 4282–4296 (1996).
Lonberg-Holm, K., Gosser, L.B. & Kauer, J.C. Early alteration of poliovirus in infected cells and its specific inhibition. J. Gen. Virol. 27, 329–342 (1975).
Fricks, C.E. & Hogle, J.M. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64, 1934–1945 (1990).
Greve, J.M. et al. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J. Virol. 65, 6015–6023 (1991).
Bostina, M., Levy, H., Filman, D.J. & Hogle, J.M. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J. Virol. 85, 776–783 (2011).
Bubeck, D. et al. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79, 7745–7755 (2005).
Levy, H.C., Bostina, M., Filman, D.J. & Hogle, J.M. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J. Virol. 84, 4426–4441 (2010).
Hendry, E. et al. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7, 1527–1538 (1999).
Belnap, D.M. et al. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74, 1342–1354 (2000).
Siebert, X. & Navaza, J. UROX 2.0: an interactive tool for fitting atomic models into electron-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 65, 651–658 (2009).
Filman, D.J. et al. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8, 1567–1579 (1989).
Grant, R.A. et al. Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design. Curr. Biol. 4, 784–797 (1994).
Smith, T.J. et al. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233, 1286–1293 (1986).
McSharry, J.J., Caliguiri, L.A. & Eggers, H.J. Inhibition of uncoating of poliovirus by arildone, a new antiviral drug. Virology 97, 307–315 (1979).
Hogle, J.M., Chow, M. & Filman, D.J. Three-dimensional structure of poliovirus at 2.9Å resolution. Science 229, 1358–1365 (1985).
Smyth, M. et al. Implications for viral uncoating from the structure of bovine enterovirus. Nat. Struct. Biol. 2, 224–231 (1995).
Foo, D.G. et al. Identification of neutralizing linear epitopes from the VP1 capsid protein of Enterovirus 71 using synthetic peptides. Virus Res. 125, 61–68 (2007).
Liu, C.C. et al. Identification and characterization of a cross-neutralization epitope of Enterovirus 71. Vaccine 29, 4362–4372 (2011).
Yang, S.L., Chou, Y.T., Wu, C.N. & Ho, M.S. Annexin II binds to capsid protein VP1 of enterovirus 71 and enhances viral infectivity. J. Virol. 85, 11809–11820 (2011).
Nishimura, Y. et al. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat. Med. 15, 794–797 (2009).
Yamayoshi, S. et al. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 15, 798–801 (2009).
Rossmann, M.G., He, Y. & Kuhn, R.J. Picornavirus-receptor interactions. Trends Microbiol. 10, 324–331 (2002).
He, Y. et al. Complexes of poliovirus serotypes with their common cellular receptor, CD155. J. Virol. 77, 4827–4835 (2003).
Xiao, C. et al. The crystal structure of coxsackievirus A21 and its interaction with ICAM-1. Structure 13, 1019–1033 (2005).
Oliveira, M.A. et al. The structure of human rhinovirus 16. Structure 1, 51–68 (1993).
Kim, S. et al. Conformational variability of a picornavirus capsid: pH-dependent structural changes of Mengo virus related to its host receptor attachment site and disassembly. Virology 175, 176–190 (1990).
McDermott, B.M. Jr., Rux, A.H., Eisenberg, R.J., Cohen, G.H. & Racaniello, V.R. Two distinct binding affinities of poliovirus for its cellular receptor. J. Biol. Chem. 275, 23089–23096 (2000).
Giranda, V.L. et al. Acid-induced structural changes in human rhinovirus 14: possible role in uncoating. Proc. Natl. Acad. Sci. USA 89, 10213–10217 (1992).
Davis, M.P. et al. Recombinant VP4 of human rhinovirus induces permeability in model membranes. J. Virol. 82, 4169–4174 (2008).
Racaniello, V.R. Early events in poliovirus infection: virus-receptor interactions. Proc. Natl. Acad. Sci. USA 93, 11378–11381 (1996).
Lin, J. et al. An externalized polypeptide partitions between two distinct sites on genome-released poliovirus particles. J. Virol. 85, 9974–9983 (2011).
Kay, B.K., Williamson, M.P. & Sudol, M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14, 231–241 (2000).
Stuart, D.I., Levine, M., Muirhead, H. & Stammers, D.K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6Å. J. Mol. Biol. 134, 109–142 (1979).
Walter, T.S. et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D Biol. Crystallogr. 61, 651–657 (2005).
Axford, D. et al. In situ macromolecular crystallography using microbeams. Acta Crystallogr. D Biol. Crystallogr. (in the press).
Borek, D., Cymborowski, M., Machius, M., Minor, W. & Otwinowski, Z. Diffraction data analysis in the presence of radiation damage. Acta Crystallogr. D Biol. Crystallogr. 66, 426–436 (2010).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 (2004).
Laskowski, R.A., Moss, D.S. & Thornton, J.M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231, 1049–1067 (1993).
Acknowledgements
We thank Sinovac Biotech Ltd. and the China National Biotech Group for providing virus samples, R. Gilbert for assistance with analytical ultracentrifugation, R. Esnouf for help with pocket analysis, J. Grimes for various help, especially with VEDA, and A. Kotecha for assistance with Diamond data collection. We also thank the Photon Factory, Japan, and the National Synchrotron Radiation Laboratory (NSRL), China. Work was supported by the National Major Project of Infectious Disease, the Ministry of Science and the Technology 973 Project (grant no. 2007CB914304). D.I.S., E.E.F. and T.S.W. are supported by the UK Medical Research Council, J.R. by the Wellcome Trust and C.P. by the Department for Environment, Food and Rural Affairs (DEFRA, UK).
Author information
Authors and Affiliations
Contributions
J.W., Z.H., W.Y. and X.S. prepared samples; X.W., W.P., X.L., Z.L., J.X., J.R., C.P., G.E., D.A., R.O., T.S.W., E.E.F. and D.I.S. performed research; W.P., X.W., Z.L., J.R., E.E.F. and D.I.S. analyzed data and, with D.J.R. and Z.R., wrote the manuscript, in discussion with J.W., Z.H., W.Y. and X.S.; all authors contributed to experimental design; Z.R. and D.I.S. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Table 1 (PDF 10558 kb)
Rights and permissions
About this article
Cite this article
Wang, X., Peng, W., Ren, J. et al. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol 19, 424–429 (2012). https://doi.org/10.1038/nsmb.2255
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2255
This article is cited by
-
Current status of hand-foot-and-mouth disease
Journal of Biomedical Science (2023)
-
Global emergence of Enterovirus 71: a systematic review
Beni-Suef University Journal of Basic and Applied Sciences (2022)
-
Transcriptome of human neuroblastoma SH-SY5Y cells in response to 2B protein of enterovirus-A71
Scientific Reports (2022)
-
Cryo-electron microscopy and image classification reveal the existence and structure of the coxsackievirus A6 virion
Communications Biology (2022)
-
Bioinformatics-based prediction of conformational epitopes for Enterovirus A71 and Coxsackievirus A16
Scientific Reports (2021)