Abstract
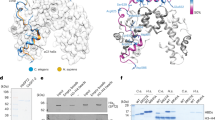
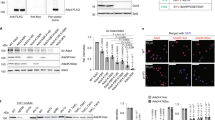
Methylation of histones is central to chromatin regulation, and thus previously unknown mechanisms regulating genome function can be revealed through the discovery of new histone methyl marks. Here we identify Set5 as the first histone H4 methyltransferase, which monomethylates the critical H4 lysine residues 5, 8 and 12 in budding yeast. Set5's enzymatic activity functions together with the global chromatin-modifying complexes COMPASS and NuA4 to regulate cell growth and stress responses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bannister, A.J. & Kouzarides, T. Cell Res. 21, 381–395 (2011).
Millar, C.B. & Grunstein, M. Nat. Rev. Mol. Cell Biol. 7, 657–666 (2006).
Garcia, B.A. et al. J. Biol. Chem. 282, 7641–7655 (2007).
Tan, M. et al. Cell 146, 1016–1028 (2011).
Sinha, H. et al. Genetics 180, 1661–1670 (2008).
Hanway, D. et al. Proc. Natl. Acad. Sci. USA 99, 10605–10610 (2002).
Teixeira, M.C., Raposo, L.R., Mira, N.P., Lourenco, A.B. & Sa-Correia, I. Appl. Environ. Microbiol. 75, 5761–5772 (2009).
Deutschbauer, A.M., Williams, R.M., Chu, A.M. & Davis, R.W. Proc. Natl. Acad. Sci. USA 99, 15530–15535 (2002).
Miller, T. et al. Proc. Natl. Acad. Sci. USA 98, 12902–12907 (2001).
Krogan, N.J. et al. J. Biol. Chem. 277, 10753–10755 (2002).
Choy, J.S., Tobe, B.T., Huh, J.H. & Kron, S.J. J. Biol. Chem. 276, 43653–43662 (2001).
Nourani, A. et al. Mol. Cell. Biol. 21, 7629–7640 (2001).
Krogan, N.J. et al. Proc. Natl. Acad. Sci. USA 101, 13513–13518 (2004).
Ma, X.J., Wu, J., Altheim, B.A., Schultz, M.C. & Grunstein, M. Proc. Natl. Acad. Sci. USA 95, 6693–6698 (1998).
Durrin, L.K., Mann, R.K., Kayne, P.S. & Grunstein, M. Cell 65, 1023–1031 (1991).
Acknowledgements
The authors are grateful to A. Morrison for critical discussions regarding the work and to members of the Gozani and Chua labs for insights. The authors also acknowledge S. Dent (The University of Texas MD Anderson Cancer Center), S. Briggs (Purdue University) and M. Cyert (Stanford University) for yeast strains and reagents. This work was supported in part by grants from the US National Institutes of Health (NIH) to O.G. (R01 GM079641) and to B.A.G. (DP2OD007447) and by a National Science Foundation CAREER award (to B.A.G.). G.M. was funded by a postdoctoral fellowship from the Ministerio de Ciencia e Innovación (Spain), and E.M.G. acknowledges support from a Cancer Biology Program postdoctoral fellowship at Stanford University. N.L.Y. was supported with an NIH F32 National Research Service Award. O.G. is the recipient of an Ellison Senior Scholar in Aging Award.
Author information
Authors and Affiliations
Contributions
E.M.G., G.M. and O.G. conceived of and designed the experiments and wrote the manuscript. G.M. and E.M.G. contributed equally to the work and conducted all biochemical and cellular experiments. MS analysis was carried out by N.L.Y. and B.A.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1 and 2 and Supplementary Methods (PDF 1963 kb)
Rights and permissions
About this article
Cite this article
Green, E., Mas, G., Young, N. et al. Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nat Struct Mol Biol 19, 361–363 (2012). https://doi.org/10.1038/nsmb.2252
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2252
This article is cited by
-
Acetyl-methyllysine marks chromatin at active transcription start sites
Nature (2023)
-
Diverse and dynamic forms of gene regulation by the S. cerevisiae histone methyltransferase Set1
Current Genetics (2023)
-
Biological function and regulation of histone 4 lysine 20 methylation in DNA damage response
Genome Instability & Disease (2022)
-
Gene repression in S. cerevisiae—looking beyond Sir-dependent gene silencing
Current Genetics (2021)
-
SET domains and stress: uncovering new functions for yeast Set4
Current Genetics (2019)