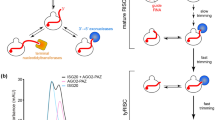
Abstract
Small RNAs, such as microRNAs and small interfering RNAs, act through Argonaute (Ago) proteins as a part of RNA-induced silencing complexes (RISCs). To make RISCs, Ago proteins bind and subsequently unwind small RNA duplexes, finally leaving one strand stably incorporated. Here we identified the N domain of human AGO2 as the initiator of duplex unwinding during RISC assembly. We discovered that a functional N domain is strictly required for small RNA duplex unwinding but not for precedent duplex loading or subsequent target cleavage. We postulate that RISC assembly is tripartite, comprising (i) RISC loading, whereby Ago undergoes conformational opening and loads a small RNA duplex, forming pre-RISC; (ii) wedging, whereby the end of the duplex is pried open through active wedging by the N domain, in preparation for unwinding; and (iii) unwinding, whereby the passenger strand is removed through slicer-dependent or slicer-independent unwinding, forming mature RISC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Hutvagner, G. & Simard, M.J. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22–32 (2008).
Carthew, R.W. & Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 (2009).
Ghildiyal, M. & Zamore, P.D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108 (2009).
Kim, V.N., Han, J. & Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139 (2009).
Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R. & Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293, 1146–1150 (2001).
Kawamata, T. & Tomari, Y. Making RISC. Trends Biochem. Sci. 35, 368–376 (2010).
Kawamata, T., Yoda, M. & Tomari, Y. Multilayer checkpoints for microRNA authenticity during RISC assembly. EMBO Rep. 12, 944–949 (2011).
Kwak, P.B., Iwasaki, S. & Tomari, Y. The microRNA pathway and cancer. Cancer Sci. 101, 2309–2315 (2010).
Matranga, C., Tomari, Y., Shin, C., Bartel, D.P. & Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123, 607–620 (2005).
Rand, T.A., Petersen, S., Du, F. & Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123, 621–629 (2005).
Miyoshi, K., Tsukumo, H., Nagami, T., Siomi, H. & Siomi, M.C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 19, 2837–2848 (2005).
Leuschner, P.J., Ameres, S.L., Kueng, S. & Martinez, J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 7, 314–320 (2006).
Kawamata, T., Seitz, H. & Tomari, Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 16, 953–960 (2009).
Yoda, M. et al. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 17, 17–23 (2010).
Iki, T. et al. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 39, 282–291 (2010).
Iwasaki, S. et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 39, 292–299 (2010).
Miyoshi, T., Takeuchi, A., Siomi, H. & Siomi, M.C. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat. Struct. Mol. Biol. 17, 1024–1026 (2010).
Johnston, M., Geoffroy, M.C., Sobala, A., Hay, R. & Hutvagner, G. HSP90 protein stabilizes unloaded Argonaute complexes and microscopic P-bodies in human cells. Mol. Biol. Cell 21, 1462–1469 (2010).
Schwarz, D.S. et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003).
Khvorova, A., Reynolds, A. & Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 (2003).
Song, J.J., Smith, S.K., Hannon, G.J. & Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437 (2004).
Yuan, Y.R. et al. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 19, 405–419 (2005).
Wang, Y., Sheng, G., Juranek, S., Tuschl, T. & Patel, D.J. Structure of the guide-strand-containing Argonaute silencing complex. Nature 456, 209–213 (2008).
Ma, J.B. et al. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434, 666–670 (2005).
Frank, F., Sonenberg, N. & Nagar, B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465, 818–822 (2010).
Boland, A., Huntzinger, E., Schmidt, S., Izaurralde, E. & Weichenrieder, O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc. Natl. Acad. Sci. USA 108, 10466–10471 (2011).
Parker, J.S., Roe, S.M. & Barford, D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434, 663–666 (2005).
Ma, J.B., Ye, K. & Patel, D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429, 318–322 (2004).
Wang, Y. et al. Structure of an Argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456, 921–926 (2008).
Wang, Y. et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461, 754–761 (2009).
Liu, J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 (2004).
Rüdel, S. et al. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 39, 2330–2343 (2011).
Liu, Y. et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 325, 750–753 (2009).
Tian, Y. et al. Multimeric assembly and biochemical characterization of the Trax-translin endonuclease complex. Nat. Struct. Mol. Biol. 18, 658–664 (2011).
Ye, X. et al. Structure of C3PO and mechanism of human RISC activation. Nat. Struct. Mol. Biol. 18, 650–657 (2011).
Parker, J.S. How to slice: snapshots of Argonaute in action. Silence 1, 3 (2010).
Nykänen, A., Haley, B. & Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107, 309–321 (2001).
Waterhouse, A.M., Procter, J.B., Martin, D.M., Clamp, M. & Barton, G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Meister, G. et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197 (2004).
Haley, B., Tang, G. & Zamore, P.D. In vitro analysis of RNA interference in Drosophila melanogaster. Methods 30, 330–336 (2003).
Kawamata, T. & Tomari, Y. Native gel analysis for RISC assembly. Methods Mol. Biol. 725, 91–105 (2011).
Acknowledgements
We acknowledge T. Tuschl (Rockefeller University) for the pIRESneo-Flag-hemagglutinin-AGO2 plasmid and S. Katsuma (The University of Tokyo) for technical assistance. We are grateful to H. Seitz, T. Kawamata and members of the Tomari laboratory for helpful discussions and suggestions and critical comments on the manuscript. This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas (“Functional machinery for non-coding RNAs”) to Y.T. P.B.K. is a Prins Bernhard Cultuurfonds Fellow, supported by the Banning-de Jong Fonds.
Author information
Authors and Affiliations
Contributions
P.B.K. conducted all experiments, Y.T. supervised the study, and P.B.K. and Y.T. wrote the manuscript, discussed the results and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1 and 2 (PDF 3536 kb)
Rights and permissions
About this article
Cite this article
Kwak, P., Tomari, Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol 19, 145–151 (2012). https://doi.org/10.1038/nsmb.2232
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2232
This article is cited by
-
Genome-wide identification, characterization and expression analysis of AGO, DCL, and RDR families in Chenopodium quinoa
Scientific Reports (2023)
-
Regulation of microRNA expression by the adaptor protein GRB2
Scientific Reports (2023)
-
Structural basis for sequence-specific recognition of guide and target strands by the Archaeoglobus fulgidus Argonaute protein
Scientific Reports (2023)
-
Functional identification of microRNA-centered complexes in C. elegans
Scientific Reports (2022)
-
Single-molecule FRET uncovers hidden conformations and dynamics of human Argonaute 2
Nature Communications (2022)