Abstract
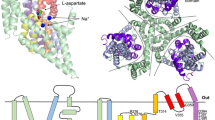
Neurotransmitter sodium symporters (NSSs) catalyze the uptake of neurotransmitters into cells, terminating neurotransmission at chemical synapses. Consistent with the role of NSSs in the central nervous system, they are implicated in multiple diseases and disorders. LeuT, from Aquifex aeolicus, is a prokaryotic ortholog of the NSS family and has contributed to our understanding of the structure, mechanism and pharmacology of NSSs. At present, however, the functional state of LeuT in crystals grown in the presence of n-octyl-β-D-glucopyranoside (β-OG) and the number of substrate binding sites are controversial issues. Here we present crystal structures of LeuT grown in DMPC-CHAPSO bicelles and demonstrate that the conformations of LeuT–substrate complexes in lipid bicelles and in β-OG detergent micelles are nearly identical. Furthermore, using crystals grown in bicelles and the substrate leucine or the substrate analog selenomethionine, we find only a single substrate molecule in the primary binding site.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Masson, J., Sagne, C., Hamon, M. & Mestikawy, S.E. Neurotransmitter transporters in the central nervous system. Pharmacol. Rev. 51, 439–464 (1999).
Amara, S.G. & Sonders, M.S. Neurotransmitter transporters as molecular targets for addictive drugs. Drug Alcohol Depend. 51, 87–96 (1998).
Gouaux, E. The molecular logic of sodium-coupled neurotransmitter transporters. Phil. Trans. R. Soc. Lond. B 364, 149–154 (2009).
Hahn, M.K. & Blakely, R.D. Monoamine transporter gene structure and polymorphisms in relation to psychiatric and other complex disorders. Pharmacogenomics J. 2, 217–235 (2002).
Richerson, G.B. & Wu, Y. Role of the GABA transporter in epilepsy. Adv. Exp. Med. Biol. 548, 76–91 (2004).
Shannon, J.R. et al. Orthostatic intolerance and tachycardia associated with norepinephrine-transporter deficiency. N. Engl. J. Med. 342, 541–549 (2000).
White, K.J., Walline, C. & Barker, E. Serotonin transporters: implications for antidepressant drug development. AAPS J. 7, E421–E433 (2005).
Dalby, N.O. Inhibition of γ-aminobutyric acid uptake: anatomy, physiology and effects against epileptic seizures. Eur. J. Pharmacol. 479, 127–137 (2003).
Krogsgaard-Larsen, P., Frolund, B. & Frydenvang, K. GABA uptake inhibitors. Design, molecular pharmacology and therapeutic aspects. Curr. Pharm. Des. 6, 1193–1209 (2000).
Yamashita, A., Singh, S.K., Kawate, T., Jin, Y. & Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223 (2005).
Singh, S.K., Piscitelli, C.L., Yamashita, A. & Gouaux, E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science 322, 1655–1661 (2008).
Singh, S.K., Yamashita, A. & Gouaux, E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature 448, 952–956 (2007).
Zhou, Z. et al. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science 317, 1390–1393 (2007).
Claxton, D.P. et al. Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:sodium symporters. Nat. Struct. Mol. Biol. 17, 822–829 (2010).
Zhao, Y. et al. Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature 474, 109–113 (2011).
Zhao, Y. et al. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature 465, 188–193 (2010).
Field, J.R., Henry, L.K. & Blakely, R.D. Transmembrane domain 6 of the human serotonin transporter contributes to an aqueously accessible binding pocket for serotonin and the psychostimulant 3,4-methylene dioxymethamphetamine. J. Biol. Chem. 285, 11270–11280 (2010).
Forrest, L.R. et al. Mechanism for alternating access in neurotransmitter transporters. Proc. Natl. Acad. Sci. USA 105, 10338–10343 (2008).
Sinning, S. et al. Binding and orientation of tricyclic antidepressants within the central substrate site of the human serotonin transporter. J. Biol. Chem. 285, 8363–8374 (2010).
Beuming, T. et al. The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nat. Neurosci. 11, 780–789 (2008).
Kniazeff, J. et al. An intracellular interaction network regulates conformational transitions in the dopamine transporter. J. Biol. Chem. 283, 17691–17701 (2008).
Kanner, B.I. & Zomot, E. Sodium-coupled neurotransmitter transporters. Chem. Rev. 108, 1654–1668 (2008).
Quick, M. et al. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc. Natl. Acad. Sci. USA 106, 5563–5568 (2009).
Shi, L., Quick, M., Zhao, Y., Weinstein, H. & Javitch, J.A. The mechanism of a neurotransmitter: sodium symporter–inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell 30, 667–677 (2008).
Piscitelli, C.L., Krishnamurthy, H. & Gouaux, E. Neurotransmitter/sodium symporter orthologue LeuT has a single high-affinity substrate site. Nature 468, 1129–1132 (2010).
Faham, S. & Bowie, J.U. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J. Mol. Biol. 316, 1–6 (2002).
Faham, S., Ujwal, R., Abramson, J. & Bowie, J.U. Chapter 5 Practical aspects of membrane proteins crystallization in bicelles. Curr. Top. Membr 63, 109–125 (2009).
Ostermeier, C. & Michel, H. Crystallization of membrane proteins. Curr. Opin. Struct. Biol. 7, 697–701 (1997).
Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 33, 491–497 (1968).
Boudker, O. & Verdon, G. Structural perspectives on secondary active transporters. Trends Pharmacol. Sci. 31, 418–426 (2010).
Smicun, Y., Campbell, S.D., Chen, M.A., Gu, H. & Rudnick, G. The role of external loop regions in serotonin transport. J. Biol. Chem. 274, 36058–36064 (1999).
Stephan, M.M., Chen, M.A., Penado, K.M. & Rudnick, G. An extracellular loop region of the serotonin transporter may be involved in the translocation mechanism. Biochemistry 36, 1322–1328 (1997).
Mitchell, S.M., Lee, E., Garcia, M.L. & Stephan, M.M. Structure and function of extracellular loop 4 of the serotonin transporter as revealed by cysteine-scanning mutagenesis. J. Biol. Chem. 279, 24089–24099 (2004).
Huber, R.E. & Criddle, R.S. The isolation and properties of β-galactosidase from Escherichia coli grown on sodium selenate. Biochim. Biophys. Acta 141, 587–599 (1967).
Hendrickson, W.A., Horton, J.R. & LeMaster, D.M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9, 1665–1672 (1990).
Mudd, S.H. & Cantoni, G.L. Selenomethionine in enzymatic transmethylations. Nature 180, 1052 (1957).
Reyes, N. & Tavoulari, S. To be, or not to be two sites: that is the question about LeuT substrate binding. J. Gen. Physiol. 138, 467–471 (2011).
Celik, L., Schiøtt, B. & Tajkhorshid, E. Substrate binding and formation of an occluded state in the leucine transporter. Biophys. J. 94, 1600–1612 (2008).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P.D. et al. Phenix: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Chen, V.B. et al. Molprobity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Collaborative Computing Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Acknowledgements
We thank P. Shaffer for crystallization and measurement of diffraction data for the LeuT–β-SeHG complex; K. Wang and A. Penmatsa for assistance with LeuT expression and purifications; R. Hibbs for suggestions on structure refinement; M. Kavanaugh and D. Claxton for comments; and L. Vaskalis for assistance with illustrations. We also thank the staff at beamlines 24-ID-E and 24-ID-C of the Advanced Photon Source, and the staff at 8.2.1 and 5.0.2 of the Advanced Light Source. This work was supported by the US National Institutes of Health. E.G. is an investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
H.W. and E.G. designed the research; H.W., J.E. and E.G. carried out the research and analyzed the data; and H.W. and E.G. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 5204 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Elferich, J. & Gouaux, E. Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context. Nat Struct Mol Biol 19, 212–219 (2012). https://doi.org/10.1038/nsmb.2215
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2215
This article is cited by
-
Mechanisms of membrane protein crystallization in ‘bicelles’
Scientific Reports (2022)
-
Transporters Through the Looking Glass: An Insight into the Mechanisms of Ion-Coupled Transport and Methods That Help Reveal Them
Journal of the Indian Institute of Science (2018)
-
Role of Histidine 547 of Human Dopamine Transporter in Molecular Interaction with HIV-1 Tat and Dopamine Uptake
Scientific Reports (2016)
-
HIV-1 transgenic rats display an increase in [3H]dopamine uptake in the prefrontal cortex and striatum
Journal of NeuroVirology (2016)
-
Structural evidence for functional lipid interactions in the betaine transporter BetP
The EMBO Journal (2013)