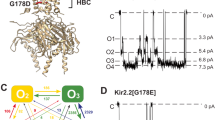
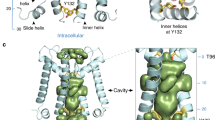
Abstract
KirBac channels are prokaryotic homologs of mammalian inwardly rectifying (Kir) potassium channels, and recent crystal structures of both Kir and KirBac channels have provided major insight into their unique structural architecture. However, all of the available structures are closed at the helix bundle crossing, and therefore the structural mechanisms that control opening of their primary activation gate remain unknown. In this study, we engineered the inner pore-lining helix (TM2) of KirBac3.1 to trap the bundle crossing in an apparently open conformation and determined the crystal structure of this mutant channel to 3.05 Å resolution. Contrary to previous speculation, this new structure suggests a mechanistic model in which rotational 'twist' of the cytoplasmic domain is coupled to opening of the bundle-crossing gate through a network of inter- and intrasubunit interactions that involve the TM2 C-linker, slide helix, G-loop and the CD loop.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hibino, H. et al. Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol. Rev. 90, 291–366 (2010).
Bichet, D., Haass, F.A. & Jan, L.Y. Merging functional studies with structures of inward-rectifier K+ channels. Nat. Rev. Neurosci. 4, 957–967 (2003).
Swartz, K.J. Towards a structural view of gating in potassium channels. Nat. Rev. Neurosci. 5, 905–916 (2004).
Kuo, A. et al. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science 300, 1922–1926 (2003).
Kuo, A., Domene, C., Johnson, L.N., Doyle, D.A. & Venien-Bryan, C. Two different conformational states of the KirBac3.1 potassium channel revealed by electron crystallography. Structure 13, 1463–1472 (2005).
Domene, C., Doyle, D.A. & Venien-Bryan, C. Modeling of an ion channel in its open conformation. Biophys J. 89, L01–L03 (2005).
Pegan, S. et al. Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat. Neurosci. 8, 279–287 (2005).
Proks, P. et al. A gating mutation at the internal mouth of the Kir6.2 pore is associated with DEND syndrome. EMBO Rep. 6, 470–475 (2005).
Proks, P., Antcliff, J.F. & Ashcroft, F.M. The ligand-sensitive gate of a potassium channel lies close to the selectivity filter. EMBO Rep. 4, 70–75 (2003).
Domene, C., Klein, M.L., Branduardi, D., Gervasio, F.L. & Parrinello, M. Conformational changes and gating at the selectivity filter of potassium channels. J. Am. Chem. Soc. 130, 9474–9480 (2008).
Tao, X., Avalos, J.L., Chen, J. & MacKinnon, R. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science 326, 1668–1674 (2009).
Nishida, M., Cadene, M., Chait, B.T. & Mackinnon, R. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 26, 4005–4015 (2007).
Clarke, O.B. et al. Domain reorientation and rotation of an intracellular assembly regulate conduction in Kir potassium channels. Cell 141, 1018–1029 (2010).
Paynter, J.J. et al. Functional complementation and genetic deletion studies of KirBac channels: activatory mutations highlight gating-sensitive domains. J. Biol. Chem. 285, 40754–40761 (2010).
Sun, S., Gan, J.H., Paynter, J.J. & Tucker, S.J. Cloning and functional characterization of a superfamily of microbial inwardly rectifying potassium channels. Physiol. Genomics 26, 1–7 (2006).
Shang, L. & Tucker, S.J. Non-equivalent role of TM2 gating hinges in heteromeric Kir4.1/Kir5.1 potassium channels. Eur. Biophys. J. 37, 165–171 (2008).
Tai, K., Haider, S., Grottesi, A. & Sansom, M.S. Ion channel gates: comparative analysis of energy barriers. Eur. Biophys. J. 38, 347–354 (2009).
Magidovich, E. & Yifrach, O. Conserved gating hinge in ligand- and voltage-dependent K+ channels. Biochemistry 43, 13242–13247 (2004).
Jin, T. et al. The βγ subunits of G proteins gate a K+ channel by pivoted bending of a transmembrane segment. Mol. Cell 10, 469–481 (2002).
Grottesi, A., Domene, C., Hall, B. & Sansom, M.S. Conformational dynamics of M2 helices in KirBac channels: helix flexibility in relation to gating via molecular dynamics simulations. Biochemistry 44, 14586–14594 (2005).
Cuello, L.G. et al. Structural basis for the coupling between activation and inactivation gates in K+ channels. Nature 466, 272–275 (2010).
Cuello, L.G., Jogini, V., Cortes, D.M. & Perozo, E. Structural mechanism of C-type inactivation in K+ channels. Nature 466, 203–208 (2010).
Uysal, S. et al. Mechanism of activation gating in the full-length KcsA K+ channel. Proc. Natl. Acad. Sci. USA 108, 11896–11899 (2011).
Stanfield, P.R. et al. A single aspartate residue is involved in both intrinsic gating and blockage by Mg2+ of the inward rectifier, IRK1. J. Physiol. (Lond.) 478, 1–6 (1994).
Zhou, Y., Morais-Cabral, J.H., Kaufman, A. & MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001).
Bernèche, S. & Roux, B. Energetics of ion conduction through the K+ channel. Nature 414, 73–77 (2001).
Singh, D.K., Rosenhouse-Dantsker, A., Nichols, C.G., Enkvetchakul, D. & Levitan, I. Direct regulation of prokaryotic Kir channel by cholesterol. J. Biol. Chem. 284, 30727–30736 (2009).
Rosenhouse-Dantsker, A., Leal-Pinto, E., Logothetis, D.E. & Levitan, I. Comparative analysis of cholesterol sensitivity of Kir channels: role of the CD loop. Channels (Austin) 4, 63–66 (2010).
Gupta, S. et al. Conformational changes during the gating of a potassium channel revealed by structural mass spectrometry. Structure 18, 839–846 (2010).
Khurana, A. et al. Forced gating motions by a substituted titratable side chain at the bundle crossing of a potassium channel. J. Biol. Chem. 286, 36686–36693 (2011).
Cordero-Morales, J.F. et al. Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 13, 311–318 (2006).
D'Avanzo, N., Cheng, W.W., Wang, S., Enkvetchakul, D. & Nichols, C.G. Lipids driving protein structure? Evolutionary adaptations in Kir channels. Channels (Austin) 4, 139–141 (2010).
Tucker, S.J. & Baukrowitz, T. How highly charged anionic lipids bind and regulate ion channels. J. Gen. Physiol. 131, 431–438 (2008).
Leal-Pinto, E. et al. Gating of a G protein-sensitive mammalian Kir3.1 prokaryotic Kir channel chimera in planar lipid bilayers. J. Biol. Chem. 285, 39790–39800 (2010).
Hansen, S.B., Tao, X. & MacKinnon, R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–498 (2011).
Taraska, J.W. & Zagotta, W.N. Structural dynamics in the gating ring of cyclic nucleotide-gated ion channels. Nat. Struct. Mol. Biol. 14, 854–860 (2007).
Schumacher, M.A., Rivard, A.F., Bachinger, H.P. & Adelman, J.P. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature 410, 1120–1124 (2001).
Long, S.B., Campbell, E.B. & Mackinnon, R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science 309, 903–908 (2005).
Enkvetchakul, D., Jeliazkova, I., Bhattacharyya, J. & Nichols, C.G. Control of inward rectifier K channel activity by lipid tethering of cytoplasmic domains. J. Gen. Physiol. 130, 329–334 (2007).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 (2004).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Smart, O.S., Wang, X., Wallace, B.A. & Sansom, M.S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996).
Humphrey, W., Dalke, A. & Schulten, K. VMD - Visual Molecular Dynamics. J. Mol. Graph. 14, 33–38 (1996).
Holst, M.J. & Saied, F. Numerical solution of the nonlinear Poisson-Boltzmann equation: Developing more robust and efficient methods. J. Comput. Chem. 16, 337–364 (1995).
Beckstein, O., Tai, K. & Sansom, M.S. Not ions alone: Barriers to ion permeation in nanopores and channels. J. Am. Chem. Soc. 126, 14694–14695 (2004).
Dolinsky, T.J. et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 (2007).
Van Der Spoel, D. et al. GROMACS: Fast, flexible and free. J. Comput. Chem. 26, 1701–1718 (2005).
Sali, A.A. & Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Acknowledgements
We thank the staff at the I24 beamline at the Diamond Light Source. This work was supported by the Biotechnology and Biological Sciences Research Council and the Wellcome Trust. R. DeZ was supported by a Marie Curie Intra-European Fellowship.
Author information
Authors and Affiliations
Contributions
S.J.T. and C.V.-B. conceived and designed the research. R. De Z. and V.N.B. expressed and crystallized the mutant protein. V.N.B., R. De Z. and L.Z. collected the diffraction data. V.N.B. and J.R.C.M. determined and refined the structure with contribution from R. De Z., V.N.B., L.Z. and M.R.S. analyzed and interpreted the structure. L.Z. conducted complementation studies. C.V.-B., M.S.P.S. and S.J.T. supervised the project. V.N.B. and S.J.T. wrote the manuscript with the help of comments from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 1440 kb)
Supplementary Movie 1
Opening of the bundle-crossing. Linear interpolation between the non-twist closed state (PDB 2WLJ) to the intermediate twist closed state (PDB 2X6C) and finally to the new, twisted S129R open conformation (PDB 3ZRS). For clarity of presentation, missing residues in the extracellular loops of 2X6C and the βL-M loop of 3ZRS were modelled in. (MOV 2508 kb)
Supplementary Movie 2
Network of Interactions. View of the network of interactions that connects the slide-helix and C-linker to the CD-loop and G-loops on the intracellular assembly. Arg137 on the C-linker and Arg-167 on the CD-loop are critical to integration of this network and may play a key role in controlling the position of the C-linker relative to the CTD. (MOV 5667 kb)
Rights and permissions
About this article
Cite this article
Bavro, V., De Zorzi, R., Schmidt, M. et al. Structure of a KirBac potassium channel with an open bundle crossing indicates a mechanism of channel gating. Nat Struct Mol Biol 19, 158–163 (2012). https://doi.org/10.1038/nsmb.2208
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2208
This article is cited by
-
A small-molecule activation mechanism that directly opens the KCNQ2 channel
Nature Chemical Biology (2024)
-
Subunit gating resulting from individual protonation events in Kir2 channels
Nature Communications (2023)
-
New Structural insights into Kir channel gating from molecular simulations, HDX-MS and functional studies
Scientific Reports (2020)
-
A constricted opening in Kir channels does not impede potassium conduction
Nature Communications (2020)
-
Coupling of a viral K+-channel with a glutamate-binding-domain highlights the modular design of ionotropic glutamate-receptors
Communications Biology (2019)