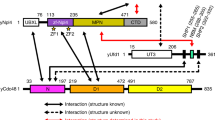
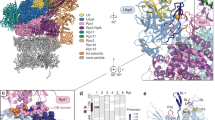
Abstract
Addition and removal of ubiquitin or ubiquitin chains to and from proteins is a tightly regulated process that contributes to cellular signaling and protein stability. Here we show that phosphorylation of the human deubiquitinase DUBA (OTUD5) at a single residue, Ser177, is both necessary and sufficient to activate the enzyme. The crystal structure of the ubiquitin aldehyde adduct of active DUBA reveals a marked cooperation between phosphorylation and substrate binding. An intricate web of interactions involving the phosphate and the C-terminal tail of ubiquitin cause DUBA to fold around its substrate, revealing why phosphorylation is essential for deubiquitinase activity. Phosphoactivation of DUBA represents an unprecedented mode of protease regulation and a clear link between two major cellular signal transduction systems: phosphorylation and ubiquitin modification.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Reyes-Turcu, F.E., Ventii, K.H. & Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 (2009).
Sowa, M.E., Bennett, E.J., Gygi, S.P. & Harper, J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009).
López-Otín, C. & Hunter, T. The regulatory crosstalk between kinases and proteases in cancer. Nat. Rev. Cancer 10, 278–292 (2010).
Kurokawa, M. & Kornbluth, S. Caspases and kinases in a death grip. Cell 138, 838–854 (2009).
Dephoure, N. et al. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 105, 10762–10767 (2008).
Kayagaki, N. et al. DUBA: a deubiquitinase that regulates type I interferon production. Science 318, 1628–1632 (2007).
Edelmann, M.J. et al. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem. J. 418, 379–390 (2009).
Komander, D. & Barford, D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem. J. 409, 77–85 (2008).
Lin, S.-C. et al. Molecular basis for the unique deubiquitinating activity of the NF-KB inhibitor A20. J. Mol. Biol. 376, 526–540 (2008).
Hurley, J.H., Lee, S. & Prag, G. Ubiquitin-binding domains. Biochem. J. 399, 361–372 (2006).
Mayya, V. et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2, ra46 (2009).
Songyang, Z. et al. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell Biol. 16, 6486–6493 (1996).
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Hu, M. et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111, 1041–1054 (2002).
Hirsch, A.K.H., Fischer, F.R. & Diederich, F. Phosphate recognition in structural biology. Angew. Chem. Int. Ed. Engl. 46, 338–352 (2007).
Taylor, S.S. & Kornev, A.P. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 36, 65–77 (2011).
Deshaies, R.J. & Joazeiro, C.A.P. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009).
Komander, D. Mechanism, specificity, and structure of the deubiquitinases. Subcell. Biochem. 54, 69–87 (2010).
Bourhis, E. et al. Reconstitution of a Frizzled8-Wnt3a-LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. J. Biol. Chem. 285, 9172–9179 (2010).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Borodovsky, A. et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9, 1149–1159 (2002).
Dong, K.C. et al. Preparation of distinct ubiquitin chain reagents of high purity and yield. Structure 19, 1053–1063 (2011).
Jones, J.W. et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. USA 107, 9771–9776 (2010).
Wang, G.G. et al. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat. Methods 3, 287–293 (2006).
Acknowledgements
We thank K.C. Dong (Genentech) for the gift of Ub-BEA, M.P. Kamps (University of California, San Diego) for the gift of ER-Hoxb8 retrovirus, T. Kitamura (University of Tokyo) for the gift of pMXs-puro, the Genentech baculovirus cloning and expression group for production of DUBA in insect cells, and the Genentech oligonucleotide synthesis and sequencing groups. Portions of this research were carried out at the Advanced Light Source, supported by the Director, Office of Science, Office of Basic Energy Sciences of the US Department of Energy, under Contract No. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Contributions
Proteins and complexes were purified by O.W.H. and J.Y.; O.W.H. carried out all biochemical studies, with advice from A.G.C., and J.Y. crystallized proteins under advice from M.A.S. J.F. and T.M. conducted the NMR analysis, with advice from M.A.S. I.B. and X.M. solved the crystal structures of apo DUBA and the ubiquitin complex, respectively, with advice from S.G.H. Q.P. and D.A. provided mass spectral data. Evaluation of DUBA phosphorylation in macrophages was done by N.K. and V.M.D. The manuscript was written by A.G.C., with contributions from O.W.H., X.M., J.Y., T.M., N.K., Q.P., S.G.H. and M.A.S.
Corresponding author
Ethics declarations
Competing interests
All authors are employees of Genentech, a member of the Roche Group.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Methods (PDF 19557 kb)
Supplementary Video 1
DUBA apo structure “morphing” into the pS177 DUBA Ub-al complex structure. (AVI 8518 kb)
Supplementary Data
Backbone 1H, 15N, and 13Cα assignments for apo DUBA OTU domain, pSer177 OTU domain, and the BEA-ubiquitin adduct of the pSer177 OTU domain. (XLS 42 kb)
Rights and permissions
About this article
Cite this article
Huang, O., Ma, X., Yin, J. et al. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat Struct Mol Biol 19, 171–175 (2012). https://doi.org/10.1038/nsmb.2206
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2206
This article is cited by
-
Phosphorylation-dependent deubiquitinase OTUD3 regulates YY1 stability and promotes colorectal cancer progression
Cell Death & Disease (2024)
-
The DUBA-SLC7A11-c-Myc axis is critical for stemness and ferroptosis
Oncogene (2023)
-
Autophagy of OTUD5 destabilizes GPX4 to confer ferroptosis-dependent kidney injury
Nature Communications (2023)
-
OTUD5 promotes innate antiviral and antitumor immunity through deubiquitinating and stabilizing STING
Cellular & Molecular Immunology (2021)
-
Deubiquitylases in developmental ubiquitin signaling and congenital diseases
Cell Death & Differentiation (2021)