Abstract
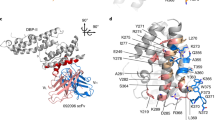
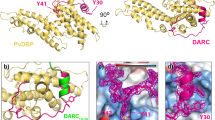
Plasmodium vivax and Plasmodium knowlesi invasion depends on the parasite Duffy-binding protein DBL domain (RII-PvDBP or RII-PkDBP) engaging the Duffy antigen receptor for chemokines (DARC) on red blood cells. Inhibition of this key interaction provides an excellent opportunity for parasite control. There are competing models for whether Plasmodium ligands engage receptors as monomers or dimers, a question whose resolution has profound implications for parasite biology and control. We report crystallographic, solution and functional studies of RII-PvDBP showing that dimerization is required for and driven by receptor engagement. This work provides a unifying framework for prior studies and accounts for the action of naturally acquired blocking antibodies and the mechanism of immune evasion. We show that dimerization is conserved in DBL-domain receptor engagement and propose that receptor-mediated ligand dimerization drives receptor affinity and specificity. Because dimerization is prevalent in signaling, our studies raise the possibility that induced dimerization may activate pathways for invasion.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Mason, S.J., Miller, L.H., Shiroishi, T., Dvorak, J.A. & McGinniss, M.H. The Duffy blood group determinants: their role in the susceptibility of human and animal erythrocytes to Plasmodium knowlesi malaria. Br. J. Haematol. 36, 327–335 (1977).
Miller, L.H., Mason, S., Clyde, D. & McGinniss, M. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295, 302–304 (1976).
Miller, L.H., Mason, S.J. & Dvorak, J.A. Erythrocytes receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science 189, 561–563 (1975).
Adams, J.H. et al. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. USA 89, 7085–7089 (1992).
Chitnis, C.E. & Miller, L. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 180, 497–506 (1994).
Sim, B.K., Chitnis, C.E., Wasniowska, K., Hadley, T.J. & Miller, L.H. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264, 1941–1944 (1994).
Adams, J.H. et al. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell 63, 141–153 (1990).
Haynes, J.D. et al. Receptor-like specificity of a Plasmodium knowlesi malarial protein that binds to Duffy antigen ligands on erythrocytes. J. Exp. Med. 167, 1873–1881 (1988).
Reed, M.B. et al. Targeted disruption of an erythrocyte binding antigen in Plasmodium falciparum is associated with a switch toward a sialic acid-independent pathway of invasion. Proc. Natl. Acad. Sci. USA 97, 7509–7514 (2000).
Gaur, D., Mayer, D.C.G. & Miller, L.H. Parasite ligand-host receptor interactions during invasion of erythrocytes by Plasmodium merozoites. Int. J. Parasitol. 34, 1413–1429 (2004).
Narum, D.L., Fuhrmann, S.R., Luu, T. & Sim, B.K.L. A novel Plasmodium falciparum erythrocyte binding protein-2 (EBP2/BAEBL) involved in erythrocyte receptor binding. Mol. Biochem. Parasitol. 119, 159–168 (2002).
Adams, J.H., Blair, P.L., Kaneko, O. & Peterson, D.S. An expanding ebl family of Plasmodium falciparum. Trends Parasitol. 17, 297–299 (2001).
Camus, D. & Hadley, T. A Plasmodium falciparum antigen that binds to host erythrocytes and merozoites. Science 230, 553–556 (1985).
Lobo, C.-A., Rodriguez, M., Reid, M. & Lustigman, S. Glycophorin C is the receptor for the Plasmodium falciparum erythrocyte binding ligand PfEBP-2 (baebl). Blood 101, 4628–4631 (2003).
Carlton, J.M. et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455, 757–763 (2008).
Chakera, A., Seeber, R.M., John, A.E., Eidne, K.A. & Greaves, D.R. The Duffy antigen/receptor for chemokines exists in an oligomeric form in living cells and functionally antagonizes CCR5 signaling through hetero-oligomerization. Mol. Pharmacol. 73, 1362–1370 (2008).
Chitnis, C.E., Chaudhuri, A., Horuk, R., Pogo, A. & Miller, L. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J. Exp. Med. 184, 1531–1536 (1996).
Choe, H. et al. Sulphated tyrosines mediate association of chemokines and Plasmodium vivax Duffy binding protein with the Duffy antigen/receptor for chemokines (DARC). Mol. Microbiol. 55, 1413–1422 (2005).
Cole-Tobian, J.L. et al. Age acquired immunity to a Plasmodium vivax invasion ligand, the Duffy binding protein. J. Infect. Dis. 186, 531–539 (2002).
Xainli, J., Adams, J.H. & King, C.L. The erythrocyte binding motif of Plasmodium vivax duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol. Biochem. Parasitol. 111, 253–260 (2000).
Xainli, J. et al. Age-dependent cellular immune responses to Plasmodium vivax Duffy binding protein in humans. J. Immunol. 169, 3200–3207 (2002).
Chootong, P. et al. Mapping epitopes of the Plasmodium vivax Duffy binding protein with naturally acquired inhibitory antibodies. Infect. Immun. 78, 1089–1095 (2010).
Gosi, P. et al. Polymorphism patterns in Duffy-binding protein among Thai Plasmodium vivax isolates. Malar. J. 7, 112 (2008).
Singh, S.K., Hora, R., Belrhali, H., Chitnis, C.E. & Sharma, A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature 439, 741–744 (2006).
VanBuskirk, K.M., Sevova, E. & Adams, J.H. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc. Natl. Acad. Sci. USA 101, 15754–15759 (2004).
Hans, D. et al. Mapping binding residues in the Plasmodium vivax domain that binds Duffy antigen during red cell invasion. Mol. Microbiol. 55, 1423–1434 (2005).
Singh, S.K. et al. Definition of structural elements in Plasmodium vivax and P. knowlesi Duffy-binding domains necessary for erythrocyte invasion. Biochem. J. 374, 193–198 (2003).
Steketee, R.W., Nahlen, B., Parise, M. & Menendez, C. The burden of malaria in pregnancy in malaria-endemic areas. Am. J. Trop. Med. Hyg. 64, 28–35 (2001).
Salanti, A. et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 49, 179–191 (2003).
Srivastava, A. et al. Full-length extracellular region of the var2CSA variant of PfEMP1 is required for specific, high-affinity binding to CSA. Proc. Natl. Acad. Sci. USA 107, 4884–4889 (2010).
Khunrae, P., Philip, J.M., Bull, D.R. & Higgins, M.K. Structural comparison of two CSPG-binding DBL domains from the VAR2CSA protein important in malaria during pregnancy. J. Mol. Biol. 393, 202–213 (2009).
Tolia, N.H., Enemark, E.J., Sim, B.K.L. & Joshua-Tor, L Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell 122, 183–193 (2005).
McHenry, A.M. & Adams, J.H. The crystal structure of P. knowlesi DBPalpha DBL domain and its implications for immune evasion. Trends Biochem. Sci. 31, 487–491 (2006).
Wilson, I.A. & Cox, N.J. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 8, 737–771 (1990).
Withers-Martinez, C. et al. Malarial EBA-175 region VI crystallographic structure reveals a KIX-like binding interface. J. Mol. Biol. 375, 773–781 (2008).
Cérède, O., Dubremetz, J.F., Bout, D. & Lebrun, M. The Toxoplasma gondii protein MIC3 requires pro-peptide cleavage and dimerization to function as adhesin. EMBO J. 21, 2526–2536 (2002).
Jewett, T.J. & Sibley, L.D. The toxoplasma proteins MIC2 and M2AP form a hexameric complex necessary for intracellular survival. J. Biol. Chem. 279, 9362–9369 (2004).
Klemm, J.D., Schreiber, S.L. & Crabtree, G.R. Dimerization as a regulatory mechanism in signal transduction. Annu. Rev. Immunol. 16, 569–592 (1998).
Farzan, M. et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96, 667–676 (1999).
Wu, B. et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071 (2010).
Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
de la Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997).
CCP4. Collaborative Computational Project No. 4. (Acta Crystallogr. D50, 760, Daresbury, UK, 1994).
Cohen, S.X. et al. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr. D Biol. Crystallogr. 60, 2222–2229 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Hura, G.L. et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat. Methods 6, 606–612 (2009).
Putnam, C.D., Hammel, M., Hura, G.L. & Tainer, J.A. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 40, 191–285 (2007).
Konarev, P.V., Volkov, V.V., Sokolova, A.V., Koch, M.H.J. & Svergun, D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 (2003).
Franke, D. & Svergun, D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009).
Svergun, D., Barberato, C. & Koch, M.H.J. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 (1995).
Kozin, M.B. & Svergun, D.I. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 (2001).
Acknowledgements
We thank S. Beverley, D. Fremont, L. Joshua-Tor, D. Goldberg, D. Sibley, J. Vogel and E. Chen for constructive comments on the manuscript, J. Adams (University of South Florida) for providing RII-PvDBP DNA and D. Goldberg (Washington University) for additional reagents. We thank T. Lohman and R. Galletto for assistance with the AUC experiments, D. Fremont and T. Brett for assistance with the ITC experiments, J. Nix and the Molecular Biology Consortium Collaborative Access Team at the Advanced Light Source for assistance in crystallographic data collection, and G. Hura, K. Dyer and the Structurally Integrated Biology for Life Sciences Team at the Advanced Light Source for assistance in SAXS data collection funded by US Department of Energy Integrated Diffraction Analysis Technologies grant contract number DE-AC02-05CH11231. The project described was supported by grant number 080792 from the US National Institute of Allergy and Infectious Disease to N.H.T., and by US National Institutes of Health Training Grant no. AI007172 and a W.M. Keck Postdoctoral Fellowship to J.D.B.
Author information
Authors and Affiliations
Contributions
J.D.B. performed functional assays, SAXS data analysis, AUC studies, ITC studies and structure analyses. J.A.Z. cloned, purified and crystallized RII-PvDBP. N.H.T. designed the study, analyzed SAXS data, collected, processed and refined X-ray data, and analyzed the structure. All authors were involved in writing the paper, discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Methods (PDF 936 kb)
Rights and permissions
About this article
Cite this article
Batchelor, J., Zahm, J. & Tolia, N. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat Struct Mol Biol 18, 908–914 (2011). https://doi.org/10.1038/nsmb.2088
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2088
This article is cited by
-
Human monoclonal antibodies inhibit invasion of transgenic Plasmodium knowlesi expressing Plasmodium vivax Duffy binding protein
Malaria Journal (2023)
-
Structural basis for DARC binding in reticulocyte invasion by Plasmodium vivax
Nature Communications (2023)
-
Design of a stabilized non-glycosylated Pfs48/45 antigen enables a potent malaria transmission-blocking nanoparticle vaccine
npj Vaccines (2023)
-
The genome of the zoonotic malaria parasite Plasmodium simium reveals adaptations to host switching
BMC Biology (2021)
-
Erythrocyte glycophorins as receptors for Plasmodium merozoites
Parasites & Vectors (2019)