Abstract
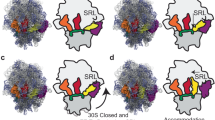
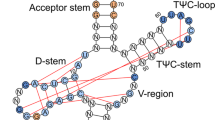
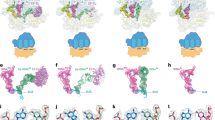
The ribosome converts genetic information into protein by selecting aminoacyl tRNAs whose anticodons base-pair to an mRNA codon. Mutations in the tRNA body can perturb this process and affect fidelity. The Hirsh suppressor is a well-studied tRNATrp harboring a G24A mutation that allows readthrough of UGA stop codons. Here we present crystal structures of the 70S ribosome complexed with EF-Tu and aminoacyl tRNA (native tRNATrp, G24A tRNATrp or the miscoding A9C tRNATrp) bound to cognate UGG or near-cognate UGA codons, determined at 3.2-Å resolution. The A9C and G24A mutations lead to miscoding by facilitating the distortion of tRNA required for decoding. A9C accomplishes this by increasing tRNA flexibility, whereas G24A allows the formation of an additional hydrogen bond that stabilizes the distortion. Our results also suggest that each native tRNA will adopt a unique conformation when delivered to the ribosome that allows accurate decoding.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hirsh, D. Tryptophan tRNA of Escherichia coli. Nature 228, 57 (1970).
Smith, D. & Yarus, M. Transfer RNA structure and coding specificity. II. A D-arm tertiary interaction that restricts coding range. J. Mol. Biol. 206, 503–511 (1989).
Smith, D. & Yarus, M. Transfer RNA structure and coding specificity. I. Evidence that a D-arm mutation reduces tRNA dissociation from the ribosome. J. Mol. Biol. 206, 489–501 (1989).
Schultz, D.W. & Yarus, M. tRNA structure and ribosomal function. I. tRNA nucleotide 27–43 mutations enhance first position wobble. J. Mol. Biol. 235, 1381–1394 (1994).
Ortiz-Meoz, R.F. & Green, R. Functional elucidation of a key contact between tRNA and the large ribosomal subunit rRNA during decoding. RNA 16, 2002–2013 (2010).
Favre, A., Buchingham, R. & Thomas, G. tRNA tertiary structure in solution as probed by the photochemically induced 8–13 cross-link. Nucleic Acids Res. 2, 1421–1431 (1975).
Cochella, L. & Green, R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308, 1178–1180 (2005).
Ogle, J.M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001).
Agris, P.F., Vendeix, F.A. & Graham, W.D. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 366, 1–13 (2007).
Pape, T., Wintermeyer, W. & Rodnina, M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 18, 3800–3807 (1999).
Parker, J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 53, 273–298 (1989).
Fersht, A.R. The hydrogen bond in molecular recognition. Trends Biochem. Sci. 12, 301–304 (1987).
Voorhees, R.M., Schmeing, T.M., Kelley, A.C. & Ramakrishnan, V. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330, 835–838 (2010).
Schmeing, T.M. et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694 (2009).
Murphy, F.V., Ramakrishnan, V., Malkiewicz, A. & Agris, P.F. The role of modifications in codon discrimination by tRNALysUUU . Nat. Struct. Mol. Biol. 11, 1186–1191 (2004).
Olejniczak, M. & Uhlenbeck, O.C. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie 88, 943–950 (2006).
Lowe, T.M. & Eddy, S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Shi, H. & Moore, P.B. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 6, 1091–1105 (2000).
Nissen, P. et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270, 1464–1472 (1995).
Li, W. et al. Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM. EMBO J. 27, 3322–3331 (2008).
Leontis, N.B. & Westhof, E. Geometric nomenclature and classification of RNA base pairs. RNA 7, 499–512 (2001).
Fahlman, R.P., Dale, T. & Uhlenbeck, O.C. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell 16, 799–805 (2004).
Gao, Y.G. et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–699 (2009).
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006).
Eisenberg, S.P., Söll, L. & Yarus, M. The purification and sequence of a temperature-sensitive tryptophan tRNA. J. Biol. Chem. 254, 5562–5566 (1979).
McCarthy, A.A. et al. A decade of user operation on the macromolecular crystallography MAD beamline ID14–4 at the ESRF. J. Synchrotron Radiat. 16, 803–812 (2009).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Acknowledgements
We thank R. Green and R. Ortiz-Meoz, Johns Hopkins University, for plasmids and bacterial strains for production of mutant tRNATrp, A. McCarthy and S. Brockhauser at ESRF ID14.4 for facilitating data collection, O. Uhlenbeck for helpful discussion and F. Murphy for scripting. This work was supported by the Medical Research Council, the Wellcome Trust, the Agouron Institute and the Louis-Jeantet Foundation. R.M.V. received support from the Gates-Cambridge scholarship. T.M.S. received support from the Human Frontier Science Program and Emmanuel College.
Author information
Authors and Affiliations
Contributions
T.M.S. and R.M.V. designed the experiments, prepared and crystallized the ribosomal complexes, collected and processed X-ray crystallography data, determined, refined and analyzed the structures, and prepared the manuscript and figures. A.C.K. purified macromolecular components, prepared and crystallized ribosomal complexes and aided with collection of X-ray crystallography data. V.R. suggested the study and provided intellectual input and supervision.
Corresponding author
Ethics declarations
Competing interests
V.R. is on the Senior Advisory Board of Rib-X Pharmaceuticals and both T.M.S. and V.R. hold stock options.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 1078 kb)
Rights and permissions
About this article
Cite this article
Schmeing, T., Voorhees, R., Kelley, A. et al. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat Struct Mol Biol 18, 432–436 (2011). https://doi.org/10.1038/nsmb.2003
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2003
This article is cited by
-
No stopping with a short-stem transfer RNA
Nature (2023)
-
2,6-Diaminopurine as a highly potent corrector of UGA nonsense mutations
Nature Communications (2020)
-
Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis
Nature Chemical Biology (2018)
-
The unique tRNASec and its role in selenocysteine biosynthesis
Amino Acids (2018)
-
Hydroxylation of a conserved tRNA modification establishes non-universal genetic code in echinoderm mitochondria
Nature Structural & Molecular Biology (2017)