Abstract
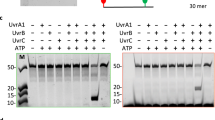
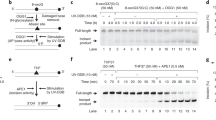
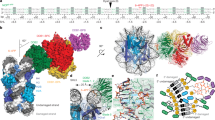
One of the primary pathways for removal of DNA damage is nucleotide excision repair (NER). In bacteria, the UvrA protein is the component of NER that locates the lesion. A notable feature of NER is its ability to act on many DNA modifications that vary in chemical structure. So far, the mechanism underlying this broad specificity has been unclear. Here, we report the first crystal structure of a UvrA protein in complex with a chemically modified oligonucleotide. The structure shows that the UvrA dimer does not contact the site of lesion directly, but rather binds the DNA regions on both sides of the modification. The DNA region harboring the modification is deformed, with the double helix bent and unwound. UvrA uses damage-induced deformations of the DNA and a less rigid structure of the modified double helix for indirect readout of the lesion.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Truglio, J.J., Croteau, D.L., Van Houten, B. & Kisker, C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 106, 233–252 (2006).
Boyce, R.P. & Howard-Flanders, P. Release of ultraviolet light-induced thymine dimers from DNA in E. coli K-12. Proc. Natl. Acad. Sci. USA 51, 293–300 (1964).
Howard-Flanders, P., Boyce, R.P. & Theriot, L. Three loci in Escherichia coli K-12 that control the excision of pyrimidine dimers and certain other mutagen products from DNA. Genetics 53, 1119–1136 (1966).
Sancar, A. & Reardon, J.T. Nucleotide excision repair in E. coli and man. Adv. Protein Chem. 69, 43–71 (2004).
Lehmann, A.R. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie 85, 1101–1111 (2003).
Cleaver, J.E., Lam, E.T. & Revet, I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat. Rev. Genet. 10, 756–768 (2009).
Sancar, A. & Rupp, W.D. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell 33, 249–260 (1983).
Machius, M., Henry, L., Palnitkar, M. & Deisenhofer, J. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc. Natl. Acad. Sci. USA 96, 11717–11722 (1999).
Nakagawa, N. et al. Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J. Biochem. 126, 986–990 (1999).
Theis, K., Chen, P.J., Skorvaga, M., Van Houten, B. & Kisker, C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 18, 6899–6907 (1999).
Truglio, J.J. et al. Structural basis for DNA recognition and processing by UvrB. Nat. Struct. Mol. Biol. 13, 360–364 (2006).
Aravind, L., Walker, D.R. & Koonin, E.V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 27, 1223–1242 (1999).
Truglio, J.J. et al. Structural insights into the first incision reaction during nucleotide excision repair. EMBO J. 24, 885–894 (2005).
Karakas, E. et al. Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad. EMBO J. 26, 613–622 (2007).
Hopfner, K.P. & Tainer, J.A. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 13, 249–255 (2003).
Linton, K.J. Structure and function of ABC transporters. Physiology (Bethesda) 22, 122–130 (2007).
Lamers, M.H. et al. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature 407, 711–717 (2000).
Obmolova, G., Ban, C., Hsieh, P. & Yang, W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407, 703–710 (2000).
Pakotiprapha, D. et al. Crystal structure of Bacillus stearothermophilus UvrA provides insight into ATP-modulated dimerization, UvrB interaction, and DNA binding. Mol. Cell 29, 122–133 (2008).
Timmins, J. et al. Structural and mutational analyses of Deinococcus radiodurans UvrA2 provide insight into DNA binding and damage recognition by UvrAs. Structure 17, 547–558 (2009).
Pakotiprapha, D., Liu, Y., Verdine, G.L. & Jeruzalmi, D. A structural model for the damage-sensing complex in bacterial nucleotide excision repair. J. Biol. Chem. 284, 12837–12844 (2009).
Oh, E.Y., Claassen, L., Thiagalingam, S., Mazur, S. & Grossman, L. ATPase activity of the UvrA and UvrAB protein complexes of the Escherichia coli UvrABC endonuclease. Nucleic Acids Res. 17, 4145–4159 (1989).
Thiagalingam, S. & Grossman, L. The multiple roles for ATP in the Escherichia coli UvrABC endonuclease-catalyzed incision reaction. J. Biol. Chem. 268, 18382–18389 (1993).
DellaVecchia, M.J. et al. Analyzing the handoff of DNA from UvrA to UvrB utilizing DNA-protein photoaffinity labeling. J. Biol. Chem. 279, 45245–45256 (2004).
Waters, T.R., Eryilmaz, J., Geddes, S. & Barrett, T.E. Damage detection by the UvrABC pathway: crystal structure of UvrB bound to fluorescein-adducted DNA. FEBS Lett. 580, 6423–6427 (2006).
Van Houten, B. & Snowden, A. Mechanism of action of the Escherichia coli UvrABC nuclease: clues to the damage recognition problem. Bioessays 15, 51–59 (1993).
Moolenaar, G.F. et al. The effect of the DNA flanking the lesion on formation of the UvrB-DNA preincision complex. Mechanism for the UvrA-mediated loading of UvrB onto a DNA damaged site. J. Biol. Chem. 275, 8038–8043 (2000).
Croteau, D.L., DellaVecchia, M.J., Perera, L. & Van Houten, B. Cooperative damage recognition by UvrA and UvrB: identification of UvrA residues that mediate DNA binding. DNA Repair (Amst.) 7, 392–404 (2008).
Bellon, S.F., Coleman, J.H. & Lippard, S.J. DNA unwinding produced by site-specific intrastrand cross-links of the antitumor drug cis-diamminedichloroplatinum(II). Biochemistry 30, 8026–8035 (1991).
Spielmann, H.P. Dynamics in psoralen-damaged DNA by 1H-detected natural abundance 13C NMR spectroscopy. Biochemistry 37, 5426–5438 (1998).
Suri, A.K., Mao, B., Amin, S., Geacintov, N.E. & Patel, D.J. Solution conformation of the (+)-trans-anti-benzo[g]chrysene-dA adduct opposite dT in a DNA duplex. J. Mol. Biol. 292, 289–307 (1999).
Kabsch, W., Sander, C. & Trifonov, E.N. The ten helical twist angles of B-DNA. Nucleic Acids Res. 10, 1097–1104 (1982).
Park, H. et al. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc. Natl. Acad. Sci. USA 99, 15965–15970 (2002).
Wiesehahn, G. & Hearst, J.E. DNA unwinding induced by photoaddition of psoralen derivatives and determination of dark-binding equilibrium constants by gel electrophoresis. Proc. Natl. Acad. Sci. USA 75, 2703–2707 (1978).
Isaacs, R.J. & Spielmann, H.P. A model for initial DNA lesion recognition by NER and MMR based on local conformational flexibility. DNA Repair (Amst.) 3, 455–464 (2004).
Oh, E.Y. & Grossman, L. The effect of Escherichia coli Uvr protein binding on the topology of supercoiled DNA. Nucleic Acids Res. 14, 8557–8571 (1986).
Mazur, S.J. & Grossman, L. Dimerization of Escherichia coli UvrA and its binding to undamaged and ultraviolet light damaged DNA. Biochemistry 30, 4432–4443 (1991).
Van Houten, B., Gamper, H., Hearst, J.E. & Sancar, A. Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. J. Biol. Chem. 263, 16553–16560 (1988).
Wagner, K., Moolenaar, G., van Noort, J. & Goosen, N. Single-molecule analysis reveals two separate DNA-binding domains in the Escherichia coli UvrA dimer. Nucleic Acids Res. 37, 1962–1972 (2009).
Croteau, D.L. et al. The C-terminal zinc finger of UvrA does not bind DNA directly but regulates damage-specific DNA binding. J. Biol. Chem. 281, 26370–26381 (2006).
Min, J.H. & Pavletich, N.P. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature 449, 570–575 (2007).
Scrima, A. et al. Structural basis of UV DNA-damage recognition by the DDB1–DDB2 complex. Cell 135, 1213–1223 (2008).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. in Methods Enzymol. 276, 307–326 (1997).
Long, F., Vagin, A.A., Young, P. & Murshudov, G.N. BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 (2008).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P.V., Grosse-Kunstleve, R.W. & Adams, P.D. The Phenix refinement framework. CCP4 Newsl. 42, 8 (2005).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Baker, N.A., Sept, D., Joseph, S., Holst, M.J. & McCammon, J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98, 10037–10041 (2001).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Lavery, R., Moakher, M., Maddocks, J.H., Petkeviciute, D. & Zakrzewska, K. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 37, 5917–5929 (2009).
Henkel, R.D., VandeBerg, J.L. & Walsh, R.A. A microassay for ATPase. Anal. Biochem. 169, 312–318 (1988).
Conlan, L.H. & Dupureur, C.M. Dissecting the metal ion dependence of DNA binding by PvuII endonuclease. Biochemistry 41, 1335–1342 (2002).
Jiang, G.H., Skorvaga, M., Croteau, D.L., Van Houten, B. & States, J.C. Robust incision of Benoz[a]pyrene-7,8-dihyrodiol-9,10-epoxide-DNA adducts by a recombinant thermoresistant interspecies combination UvrABC endonuclease system. Biochemistry 45, 7834–7843 (2006).
Acknowledgements
We thank T. Sixma and J. Bujnicki for critical reading of the manuscript, M. Rychlik, M. Cybulska and J. Dyttus for excellent technical assistance, the staff of beamline 23-2 at European Synchrotron Radiation Facility (ESRF) and beamline 14-1 at Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY) for assistance with data collection, K. Górecka and M. Figiel for their help with data collection and P. Afonine and S. Ramón Maiques for help with structure refinement. This work was supported by a European Molecular Biology Organization Installation Grant to M.N. The access to ESRF was financed by the Polish Ministry of Science and Higher Education (project no. ESRF/73/2006). The research leading to these results has also received funding from the European Community's Seventh Framework Program under grant agreement no. 226716.
Author information
Authors and Affiliations
Contributions
M.J. purified Tm-UvrA, obtained its complex crystals and carried out the biochemical experiments. K.S. and E.N. carried out the biochemical experiments. A.T. carried out the initial expression and purification studies. M.N. collected the data. M.J., E.N. and M.N. refined and analyzed the structure. M.N. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Table 1 and Supplementary Methods (PDF 1911 kb)
Supplementary Video 1
Morphing between the DNA structure observed in the Tm-UvrA–DNA complex and the model of ideal B-form DNA to visualize the deformations of the DNA. The thymine residue with fluorescein modification is shown in orange. (MOV 2288 kb)
Supplementary Video 2
Tm-UvrA surface representation with morphing of the DNA structure as in Supplementary Video 1. Protein surface was added to visualize its complementarity with deformed DNA conformation. (MOV 3755 kb)
Rights and permissions
About this article
Cite this article
Jaciuk, M., Nowak, E., Skowronek, K. et al. Structure of UvrA nucleotide excision repair protein in complex with modified DNA. Nat Struct Mol Biol 18, 191–197 (2011). https://doi.org/10.1038/nsmb.1973
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1973
This article is cited by
-
Structural and functional analyses of the echinomycin resistance conferring protein Ecm16 from Streptomyces lasalocidi
Scientific Reports (2023)
-
In vitro reconstitution of an efficient nucleotide excision repair system using mesophilic enzymes from Deinococcus radiodurans
Communications Biology (2022)
-
The mechanisms underlying antigenic variation and maintenance of genomic integrity in Mycoplasma pneumoniae and Mycoplasma genitalium
Archives of Microbiology (2021)
-
Single-molecule imaging reveals molecular coupling between transcription and DNA repair machinery in live cells
Nature Communications (2020)
-
Substrate cross-feeding affects the speed and trajectory of molecular evolution within a synthetic microbial assemblage
BMC Evolutionary Biology (2019)