Abstract
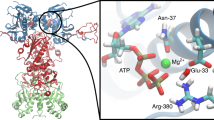
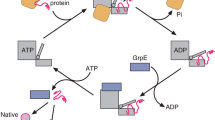
The endoplasmic reticulum is the site of folding, assembly and quality control for proteins of the secretory pathway. The ATP-regulated Hsp70 chaperone BiP (heavy chain–binding protein), together with cochaperones, has important roles in all of these processes. The functional cycle of Hsp70s is determined by conformational transitions that are required for substrate binding and release. Here, we used the intrinsically disordered CH1 domain of antibodies as an authentic substrate protein and analyzed the conformational cycle of BiP by single-molecule and ensemble Förster resonance energy transfer (FRET) measurements. Nucleotide binding resulted in concerted domain movements of BiP. Conformational transitions of the lid domain allowed BiP to discriminate between peptide and protein substrates. A major BiP cochaperone in antibody folding, ERdj3, modulated the conformational space of BiP in a nucleotide-dependent manner, placing the lid subdomain in an open, protein-accepting state.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Chen, Y. et al. SPD—a web-based secreted protein database. Nucleic Acids Res. 33, D169–D173 (2005).
Hebert, D.N. & Molinari, M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 87, 1377–1408 (2007).
Munro, S. & Pelham, H.R. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell 46, 291–300 (1986).
Karlin, S. & Brocchieri, L. Heat shock protein 70 family: multiple sequence comparisons, function, and evolution. J. Mol. Evol. 47, 565–577 (1998).
Haas, I.G. & Wabl, M. Immunoglobulin heavy chain binding protein. Nature 306, 387–389 (1983).
Weitzmann, A., Baldes, C., Dudek, J. & Zimmermann, R. The heat shock protein 70 molecular chaperone network in the pancreatic endoplasmic reticulum—a quantitative approach. FEBS J. 274, 5175–5187 (2007).
Kozutsumi, Y., Segal, M., Normington, K., Gething, M.J. & Sambrook, J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332, 462–464 (1988).
Kassenbrock, C.K., Garcia, P.D., Walter, P. & Kelly, R.B. Heavy-chain binding protein recognizes aberrant polypeptides translocated in vitro. Nature 333, 90–93 (1988).
Alder, N.N., Shen, Y., Brodsky, J.L., Hendershot, L.M. & Johnson, A.E. The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol. 168, 389–399 (2005).
Dudek, J. et al. ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat. Struct. Mol. Biol. 12, 1008–1014 (2005).
Otero, J.H., Lizak, B. & Hendershot, L.M. Life and death of a BiP substrate. Semin. Cell Dev. Biol. 21, 472–478 (2009).
Vembar, S.S. & Brodsky, J.L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944–957 (2008).
Bole, D.G., Hendershot, L.M. & Kearney, J.F. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J. Cell Biol. 102, 1558–1566 (1986).
Feige, M.J. et al. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol. Cell 34, 569–579 (2009).
Lee, Y.K., Brewer, J.W., Hellman, R. & Hendershot, L.M. BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol. Biol. Cell 10, 2209–2219 (1999).
Blond-Elguindi, S. et al. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728 (1993).
Knarr, G., Gething, M.J., Modrow, S. & Buchner, J. BiP binding sequences in antibodies. J. Biol. Chem. 270, 27589–27594 (1995).
Gething, M.J. et al. Binding sites for Hsp70 molecular chaperones in natural proteins. Cold Spring Harb. Symp. Quant. Biol. 60, 417–428 (1995).
Knarr, G., Modrow, S., Todd, A., Gething, M.J. & Buchner, J. BiP-binding sequences in HIV gp160. Implications for the binding specificity of bip. J. Biol. Chem. 274, 29850–29857 (1999).
Rüdiger, S., Germeroth, L., Schneider-Mergener, J. & Bukau, B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 (1997).
Bukau, B., Weissman, J. & Horwich, A. Molecular chaperones and protein quality control. Cell 125, 443–451 (2006).
Zhu, X. et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 (1996).
Swain, J.F. et al. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell 26, 27–39 (2007).
Bertelsen, E.B., Chang, L., Gestwicki, J.E. & Zuiderweg, E.R. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. USA 106, 8471–8476 (2009).
Vogel, M., Mayer, M.P. & Bukau, B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J. Biol. Chem. 281, 38705–38711 (2006).
Goloubinoff, P. & De Los, R.P. The mechanism of Hsp70 chaperones: (entropic) pulling the models together. Trends Biochem. Sci. 32, 372–380 (2007).
Jiang, J., Lafer, E.M. & Sousa, R. Crystallization of a functionally intact Hsc70 chaperone. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 39–43 (2006).
Mapa, K. et al. The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol. Cell 38, 89–100 (2010).
Woo, H.J., Jiang, J., Lafer, E.M. & Sousa, R. ATP-induced conformational changes in Hsp70: molecular dynamics and experimental validation of an in silico predicted conformation. Biochemistry 48, 11470–11477 (2009).
Craig, E.A., Huang, P., Aron, R. & Andrew, A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 156, 1–21 (2006).
Kampinga, H.H. & Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 (2010).
Meunier, L., Usherwood, Y.K., Chung, K.T. & Hendershot, L.M. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13, 4456–4469 (2002).
Jin, Y., Zhuang, M. & Hendershot, L.M. ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochemistry 48, 41–49 (2009).
Jin, Y., Awad, W., Petrova, K. & Hendershot, L.M. Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J. 27, 2873–2882 (2008).
Shen, Y. & Hendershot, L.M. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol. Biol. Cell 16, 40–50 (2005).
Bies, C. et al. Characterization of pancreatic ERj3p, a homolog of yeast DnaJ-like protein Scj1p. Biol. Chem. 385, 389–395 (2004).
Vembar, S.S., Jonikas, M.C., Hendershot, L.M., Weissman, J.S. & Brodsky, J.L. J domain co-chaperone specificity defines the role of BIP during protein translocation. J. Biol. Chem. 285, 22484–94 (2010).
Vanhove, M., Usherwood, Y.K. & Hendershot, L.M. Unassembled Ig heavy chains do not cycle from BiP in vivo but require light chains to trigger their release. Immunity 15, 105–114 (2001).
Knarr, G., Kies, U., Bell, S., Mayer, M. & Buchner, J. Interaction of the chaperone BiP with an antibody domain: implications for the chaperone cycle. J. Mol. Biol. 318, 611–620 (2002).
Wei, J., Gaut, J.R. & Hendershot, L.M. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J. Biol. Chem. 270, 26677–26682 (1995).
Antonik, M., Felekyan, S., Gaiduk, A. & Seidel, C.A. Separating structural heterogeneities from stochastic variations in fluorescence resonance energy transfer distributions via photon distribution analysis. J. Phys. Chem. B 110, 6970–6978 (2006).
Kalinin, S., Felekyan, S., Antonik, M. & Seidel, C.A. Probability distribution analysis of single-molecule fluorescence anisotropy and resonance energy transfer. J. Phys. Chem. B 111, 10253–10262 (2007).
Wei, J. & Hendershot, L.M. Characterization of the nucleotide binding properties and ATPase activity of recombinant hamster BiP purified from bacteria. J. Biol. Chem. 270, 26670–26676 (1995).
Jiang, J., Prasad, K., Lafer, E.M. & Sousa, R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol. Cell 20, 513–524 (2005).
Strub, A., Zufall, N. & Voos, W. The putative helical lid of the Hsp70 peptide-binding domain is required for efficient preprotein translocation into mitochondria. J. Mol. Biol. 334, 1087–1099 (2003).
Tokunaga, M., Kato, S., Kawamura-Watabe, A., Tanaka, R. & Tokunaga, H. Characterization of deletion mutations in the carboxy-terminal peptide-binding domain of the Kar2 protein in Saccharomyces cerevisiae. Yeast 14, 1285–1295 (1998).
Schlecht, R., Erbse, A.H., Bukau, B. & Mayer, M.P . Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. & Mol. Biol. (in the press).
Shen, Y., Meunier, L. & Hendershot, L.M. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 277, 15947–15956 (2002).
Vembar, S.S., Jin, Y., Brodsky, J.L. & Hendershot, L.M. The mammalian Hsp40 ERdj3 requires its Hsp70 interaction and substrate-binding properties to complement various yeast Hsp40-dependent functions. J. Biol. Chem. 284, 32462–32471 (2009).
Rodriguez, F. . et al. Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones. Mol. Cell 32, 347–358 (2008).
Mayer, M.P., Rudiger, S. & Bukau, B. Molecular basis for interactions of the DnaK chaperone with substrates. Biol. Chem. 381, 877–885 (2000).
Liu, Q. & Hendrickson, W.A. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131, 106–120 (2007).
Nørby, J.G. Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 156, 116–119 (1988).
Müller, B.K., Zaychikov, E., Brauchle, C. & Lamb, D.C. Pulsed interleaved excitation. Biophys. J. 89, 3508–3522 (2005).
Eggeling, C. et al. Data registration and selective single-molecule analysis using multi-parameter fluorescence detection. J. Biotechnol. 86, 163–180 (2001).
Kapanidis, A.N. et al. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. USA 101, 8936–8941 (2004).
Lee, N.K. et al. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys. J. 88, 2939–2953 (2005).
Schäfer, H., Nau, K., Sickmann, A., Erdmann, R. & Meyer, H.E. Identification of peroxisomal membrane proteins of Saccharomyces cerevisiae by mass spectrometry. Electrophoresis 22, 2955–2968 (2001).
Acknowledgements
We thank H. Krause for performing mass spectrometry experiments and V. Kudryavtsev for valuable discussions regarding analysis of the spFRET data. Funding of M.M. and M.J.F. by the Studienstiftung des deutschen Volkes, of M.H. by the International Doctorate Program NanoBioTechnology (IDK-NBT), of D.B. by the International Graduate School of Science and Engineering (IGSSE) and of J.B. by the SFB 749, the Fonds der chemischen Industrie and the Bayerische Forschungsstiftung is gratefully acknowledged. D.C.L. wishes to thank the Deutsche Forschungsgemeinschaft (SFB 749), the Center for Nano Science, the Ludwig-Maximilians-Universität Munich (LMUInnovativ BioImaging Network) and the Nanosystems Initative Munich (NIM) for financial support. The authors thank L. Hendershot for discussions and helpful comments on the manuscript.
Author information
Authors and Affiliations
Contributions
M.M., M.H., M.J.F., D.C.L. and J.B. designed the study and wrote the paper. M.M., M.J.F. and D.B. performed ensemble experiments and M.H. performed single molecule experiments. M.M., M.H., M.J.F. and D.C.L. analyzed data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–3 and Supplementary Methods (PDF 624 kb)
Rights and permissions
About this article
Cite this article
Marcinowski, M., Höller, M., Feige, M. et al. Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat Struct Mol Biol 18, 150–158 (2011). https://doi.org/10.1038/nsmb.1970
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1970
This article is cited by
-
Structures of a deAMPylation complex rationalise the switch between antagonistic catalytic activities of FICD
Nature Communications (2021)
-
Specificity of AMPylation of the human chaperone BiP is mediated by TPR motifs of FICD
Nature Communications (2021)
-
Urinary ERdj3 and mesencephalic astrocyte-derived neutrophic factor identify endoplasmic reticulum stress in glomerular disease
Laboratory Investigation (2020)
-
The Hsp70 chaperone network
Nature Reviews Molecular Cell Biology (2019)
-
MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP
Nature Communications (2019)