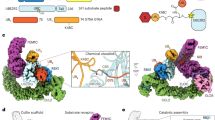
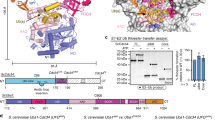
Abstract
The N-end rule pathway is a regulated proteolytic system that targets proteins containing destabilizing N-terminal residues (N-degrons) for ubiquitination and proteasomal degradation in eukaryotes. The N-degrons of type 1 substrates contain an N-terminal basic residue that is recognized by the UBR box domain of the E3 ubiquitin ligase UBR1. We describe structures of the UBR box of Saccharomyces cerevisiae UBR1 alone and in complex with N-degron peptides, including that of the cohesin subunit Scc1, which is cleaved and targeted for degradation at the metaphase-anaphase transition. The structures reveal a previously unknown protein fold that is stabilized by a novel binuclear zinc center. N-terminal arginine, lysine or histidine side chains of the N-degron are coordinated in a multispecific binding pocket. Unexpectedly, the structures together with our in vitro biochemical and in vivo pulse-chase analyses reveal a previously unknown modulation of binding specificity by the residue at position 2 of the N-degron.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bachmair, A., Finley, D. & Varshavsky, A. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 (1986).
Varshavsky, A. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93, 12142–12149 (1996).
Tobias, J.W., Shrader, T.E., Rocap, G. & Varshavsky, A. The N-end rule in bacteria. Science 254, 1374–1377 (1991).
Bachmair, A. & Varshavsky, A. The degradation signal in a short-lived protein. Cell 56, 1019–1032 (1989).
Rao, H., Uhlmann, F., Nasmyth, K. & Varshavsky, A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410, 955–959 (2001).
Turner, G.C., Du, F. & Varshavsky, A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405, 579–583 (2000).
An, J.Y. et al. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 103, 6212–6217 (2006).
Zenker, M. et al. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome). Nat. Genet. 37, 1345–1350 (2005).
Ditzel, M. et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat. Cell Biol. 5, 467–473 (2003).
Hu, R.G., Wang, H., Xia, Z. & Varshavsky, A. The N-end rule pathway is a sensor of heme. Proc. Natl. Acad. Sci. USA 105, 76–81 (2008).
Hu, R.G. et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986 (2005).
Lee, M.J. et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 102, 15030–15035 (2005).
Holman, T.J. et al. The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. USA 106, 4549–4554 (2009).
Graciet, E. et al. The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc. Natl. Acad. Sci. USA 106, 13618–13623 (2009).
Hwang, C.S., Shemorry, A. & Varshavsky, A. Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkylguanine transferase. Proc. Natl. Acad. Sci. USA 106, 2142–2147 (2009).
Tasaki, T. et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 25, 7120–7136 (2005).
Tasaki, T. et al. The substrate recognition domains of the N-end rule pathway. J. Biol. Chem. 284, 1884–1895 (2009).
Lupas, A.N. & Koretke, K.K. Bioinformatic analysis of ClpS, a protein module involved in prokaryotic and eukaryotic protein degradation. J. Struct. Biol. 141, 77–83 (2003).
Dougan, D.A., Reid, B.G., Horwich, A.L. & Bukau, B. ClpS, a substrate modulator of the ClpAP machine. Mol. Cell 9, 673–683 (2002).
Baker, R.T. & Varshavsky, A. Yeast N-terminal amidase. A new enzyme and component of the N-end rule pathway. J. Biol. Chem. 270, 12065–12074 (1995).
Kwon, Y.T. et al. Altered activity, social behavior, and spatial memory in mice lacking the NTAN1p amidase and the asparagine branch of the N-end rule pathway. Mol. Cell. Biol. 20, 4135–4148 (2000).
Bartel, B., Wunning, I. & Varshavsky, A. The recognition component of the N-end rule pathway. EMBO J. 9, 3179–3189 (1990).
Holm, L., Kaariainen, S., Rosenstrom, P. & Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008).
Suhrer, S.J., Wiederstein, M., Gruber, M. & Sippl, M.J. COPS–a novel workbench for explorations in fold space. Nucleic Acids Res. 37, W539–W544 (2009).
Marmorstein, R., Carey, M., Ptashne, M. & Harrison, S.C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature 356, 408–414 (1992).
Baleja, J.D., Marmorstein, R., Harrison, S.C. & Wagner, G. Solution structure of the DNA-binding domain of Cd2–GAL4 from S. cerevisiae. Nature 356, 450–453 (1992).
Xia, Z. et al. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J. Biol. Chem. 283, 24011–24028 (2008).
Wang, K.H., Roman-Hernandez, G., Grant, R.A., Sauer, R.T. & Baker, T.A. The molecular basis of N-end rule recognition. Mol. Cell 32, 406–414 (2008).
Roman-Hernandez, G., Grant, R.A., Sauer, R.T. & Baker, T.A. Molecular basis of substrate selection by the N-end rule adaptor protein ClpS. Proc. Natl. Acad. Sci. USA 106, 8888–8893 (2009).
Schuenemann, V.J. et al. Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS. EMBO Rep. 10, 508–514 (2009).
Wang, H., Piatkov, K.I., Brower, C.S. & Varshavsky, A. Glutamine-specific N-terminal amidase, a component of the N-end rule pathway. Mol. Cell 34, 686–695 (2009).
Du, F., Navarro-Garcia, F., Xia, Z., Tasaki, T. & Varshavsky, A. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. USA 99, 14110–14115 (2002).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000).
Lamzin, V.S. & Wilson, K.S. Automated refinement of protein models. Acta Crystallogr. D Biol. Crystallogr. 49, 129–147 (1993).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Winn, M.D., Murshudov, G.N. & Papiz, M.Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 (2003).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Laskowski, R.A., Moss, D.S. & Thornton, J.M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231, 1049–1067 (1993).
Ghislain, M., Dohmen, R.J., Levy, F. & Varshavsky, A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15, 4884–4899 (1996).
Acknowledgements
We thank the staff at the 4A and 6B beamlines, Pohang Accelerator Laboratory, Korea, and the NW12 beamline, Photon Factory, Japan, for help with the data collection. We also thank Y.T. Kwon (University of Pittsburgh) for providing us with unpublished data of UBR fragments and A. Varshavsky (California Institute of Technology) for yeast strains and plasmids used for the in vivo study. This work was supported by a Basic Science Research Program through the National Research Foundation of Korea (NRF 2007-0055395), the 21C Frontier Functional Proteomics Project (FPR08B2-270), World-Class University Project (R33-10108), the Plant Signaling Network Research Center and the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A092006).
Author information
Authors and Affiliations
Contributions
W.S.C., B.-C.J., M.-R.L. and H.K.S. performed structural studies; W.S.C. and B.-C.J. performed biochemical studies; Y.J.J. and J.K. performed in vivo assay; W.S.C., M.J.E. & H.K.S. designed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Table 1 (PDF 2204 kb)
Rights and permissions
About this article
Cite this article
Choi, W., Jeong, BC., Joo, Y. et al. Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat Struct Mol Biol 17, 1175–1181 (2010). https://doi.org/10.1038/nsmb.1907
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1907
This article is cited by
-
CRL2ZER1/ZYG11B recognizes small N-terminal residues for degradation
Nature Communications (2022)
-
Molecular basis for ubiquitin ligase CRL2FEM1C-mediated recognition of C-degron
Nature Chemical Biology (2021)
-
Structural insights into Ubr1-mediated N-degron polyubiquitination
Nature (2021)
-
Ubr1-mediated ubiquitylation orchestrates asexual development, polar growth, and virulence-related cellular events in Beauveria bassiana
Applied Microbiology and Biotechnology (2021)
-
R-catcher, a potent molecular tool to unveil the arginylome
Cellular and Molecular Life Sciences (2021)