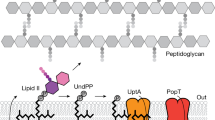
Abstract
Teichoic acid polymers are composed of polyol-phosphate units and form a major component of Gram-positive bacterial cell walls. These anionic compounds perform a multitude of important roles in bacteria and are synthesized by monotopic membrane proteins of the TagF polymerase family. We have determined the structure of Staphylococcus epidermidis TagF to 2.7-Å resolution from a construct that includes both the membrane-targeting region and the glycerol-phosphate polymerase domains. TagF possesses a helical region for interaction with the lipid bilayer, placing the active site at a suitable distance for access to the membrane-bound substrate. Characterization of active-site residue variants and analysis of a CDP-glycerol substrate complex suggest a mechanism for polymer synthesis. With the importance of teichoic acid in Gram-positive physiology, this elucidation of the molecular details of TagF function provides a critical new target in the development of novel anti-infectives.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Armstrong, J.J. et al. Composition of teichoic acids from a number of bacterial walls. Nature 184, 247–248 (1959).
Hancock, I.C. Bacterial cell surface carbohydrates: structure and assembly. Biochem. Soc. Trans. 25, 183–187 (1997).
Weidenmaier, C. & Peschel, A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6, 276–287 (2008).
Kohler, T., Weidenmaier, C. & Peschel, A. Wall teichoic acid protects Staphylococcus aureus against antimicrobial fatty acids from human skin. J. Bacteriol. 191, 4482–4484 (2009).
Oku, Y. et al. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J. Bacteriol. 191, 141–151 (2009).
Schirner, K., Marles-Wright, J., Lewis, R.J. & Errington, J. Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 28, 830–842 (2009).
D'Elia, M.A., Millar, K.E., Beveridge, T.J. & Brown, E.D. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J. Bacteriol. 188, 8313–8316 (2006).
D'Elia, M.A. et al. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 188, 4183–4189 (2006).
von Eiff, C., Heilmann, C. & Peters, G. New aspects in the molecular basis of polymer-associated infections due to staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 18, 843–846 (1999).
Xiang, Y. et al. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol. Cell 34, 375–386 (2009).
Ward, J.B. Teichoic and teichuronic acids: biosynthesis, assembly, and location. Microbiol. Rev. 45, 211–243 (1981).
Lu, D. et al. Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. Proc. Natl. Acad. Sci. USA 106, 1584–1589 (2009).
Sewell, E.W., Pereira, M.P. & Brown, E.D. The wall teichoic acid polymerase TagF is non-processive in vitro and amenable to study using steady state kinetic analysis. J. Biol. Chem. 284, 21132–21138 (2009).
Brown, S., Zhang, Y.H. & Walker, S. A revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem. Biol. 15, 12–21 (2008).
Fitzgerald, S.N. & Foster, T.J. Molecular analysis of the tagF gene, encoding CDP-Glycerol:Poly(glycerophosphate) glycerophosphotransferase of Staphylococcus epidermidis ATCC 14990. J. Bacteriol. 182, 1046–1052 (2000).
Cantarel, B.L. et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 (2009).
Bourne, Y. & Henrissat, B. Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr. Opin. Struct. Biol. 11, 593–600 (2001).
Wrabl, J.O. & Grishin, N.V. Homology between O-linked GlcNAc transferases and proteins of the glycogen phosphorylase superfamily. J. Mol. Biol. 314, 365–374 (2001).
Holm, L. & Sander, C. Protein folds and families: sequence and structure alignments. Nucleic Acids Res. 27, 244–247 (1999).
Formstone, A., Carballido-Lopez, R., Noirot, P., Errington, J. & Scheffers, D.J. Localization and interactions of teichoic acid synthetic enzymes in Bacillus subtilis. J. Bacteriol. 190, 1812–1821 (2008).
Schertzer, J.W. & Brown, E.D. Use of CDP-glycerol as an alternate acceptor for the teichoic acid polymerase reveals that membrane association regulates polymer length. J. Bacteriol. 190, 6940–6947 (2008).
Bhavsar, A.P., D′Elia, M.A., Sahakian, T.D. & Brown, E.D. The amino terminus of Bacillus subtilis TagB possesses separable localization and functional properties. J. Bacteriol. 189, 6816–6823 (2007).
Bracey, M.H., Hanson, M.A., Masuda, K.R., Stevens, R.C. & Cravatt, B.F. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science 298, 1793–1796 (2002).
Schertzer, J.W., Bhavsar, A.P. & Brown, E.D. Two conserved histidine residues are critical to the function of the TagF-like family of enzymes. J. Biol. Chem. 280, 36683–36690 (2005).
Ha, S., Walker, D., Shi, Y. & Walker, S. The 1.9 Å crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci. 9, 1045–1052 (2000).
Bolam, D.N. et al. The crystal structure of two macrolide glycosyltransferases provides a blueprint for host cell antibiotic immunity. Proc. Natl. Acad. Sci. USA 104, 5336–5341 (2007).
Ni, L. et al. Cytidine 5′-monophosphate (CMP)-induced structural changes in a multifunctional sialyltransferase from Pasteurella multocida. Biochemistry 45, 2139–2148 (2006).
Lairson, L.L., Henrissat, B., Davies, G.J. & Withers, S.G. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 (2008).
Campbell, R.E., Mosimann, S.C., Tanner, M.E. & Strynadka, N.C. The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases. Biochemistry 39, 14993–15001 (2000).
Buschiazzo, A. et al. Crystal structure of glycogen synthase: homologous enzymes catalyze glycogen synthesis and degradation. EMBO J. 23, 3196–3205 (2004).
Honeyman, A.L. & Stewart, G.C. The nucleotide sequence of the rodC operon of Bacillus subtilis. Mol. Microbiol. 3, 1257–1268 (1989).
Lomize, A.L., Pogozheva, I.D., Lomize, M.A. & Mosberg, H.I. The role of hydrophobic interactions in positioning of peripheral proteins in membranes. BMC Struct. Biol. 7, 44 (2007).
Barreras, M., Salinas, S.R., Abdian, P.L., Kampel, M.A. & Ielpi, L. Structure and mechanism of GumK, a membrane-associated glucuronosyltransferase. J. Biol. Chem. 283, 25027–25035 (2008).
Guerin, M.E. et al. Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J. Biol. Chem. 282, 20705–20714 (2007).
Golovin, A. & Henrick, K. MSDmotif: exploring protein sites and motifs. BMC Bioinformatics 9, 312 (2008).
Ni, L. et al. Crystal structures of Pasteurella multocida sialyltransferase complexes with acceptor and donor analogues reveal substrate binding sites and catalytic mechanism. Biochemistry 46, 6288–6298 (2007).
Shiraki, K., Norioka, S., Li, S., Yokota, K. & Sakiyama, F. Electrostatic role of aromatic ring stacking in the pH-sensitive modulation of a chymotrypsin-type serine protease, Achromobacter protease I. Eur. J. Biochem. 269, 4152–4158 (2002).
Burger, M.M. & Glaser, L. The synthesis of teichoic acids. I. Polyglycerophosphate. J. Biol. Chem. 239, 3168–3177 (1964).
Mauck, J. & Glaser, L. An acceptor-dependent polyglycerolphosphate polymerase. Proc. Natl. Acad. Sci. USA 69, 2386–2390 (1972).
Wickham, J.R., Halye, J.L., Kashtanov, S., Khandogin, J. & Rice, C.V. Revisiting magnesium chelation by teichoic acid with phosphorus solid-state NMR and theoretical calculations. J. Phys. Chem. B 113, 2177–2183 (2009).
Pereira, M.P. et al. The wall teichoic acid polymerase TagF efficiently synthesizes poly(glycerol phosphate) on the TagB product lipid III. Chem. Bio. Chem 9, 1385–1390 (2008).
Schertzer, J.W. & Brown, E.D. Purified, recombinant TagF protein from Bacillus subtilis 168 catalyzes the polymerization of glycerol phosphate onto a membrane acceptor in vitro. J. Biol. Chem. 278, 18002–18007 (2003).
Ramirez, M. & Tomasz, A. Molecular characterization of the complete 23F capsular polysaccharide locus of Streptococcus pneumoniae. J. Bacteriol. 180, 5273–5278 (1998).
Baker, N.A., Sept, D., Joseph, S., Holst, M.J. & McCammon, J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98, 10037–10041 (2001).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Collaborative Computation Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
Zwart, P.H. et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 (2008).
Terwilliger, T.C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Badurina, D.S., Zolli-Juran, M. & Brown, E.D. CTP:glycerol 3-phosphate cytidylyltransferase (TarD) from Staphylococcus aureus catalyzes the cytidylyl transfer via an ordered Bi-Bi reaction mechanism with micromolar Km values. Biochim. Biophys. Acta 1646, 196–206 (2003).
Acknowledgements
We thank G. Olovsson for maintenance of X-ray facilities. This work was made possible by operating grants from the Canadian Institutes of Health Research and Howard Hughes Medical Institute and infrastructure grants from the Michael Smith Foundation of Health Research and the Canadian Foundation of Innovation (to N.C.J.S.). N.C.J.S. is a Howard Hughes Medical Institute international research scholar and Michael Smith Foundation for Health Research Senior Scholar. E.D.B. was supported by a salary award from the Canada Research Chairs program and operating funds from the Canadian Institutes of Health Research (MOP-15496). We are grateful for beam time and assistance at the Advanced Light Source and Canadian Light Source.
Author information
Authors and Affiliations
Contributions
A.L.L. was responsible for the cloning, purification, crystallization and all X-ray and structural analyses; E.W.S. performed and analyzed kinetic assays; L.Y.-C.L. designed and tested the heparin purification procedure; T.S. contributed to crystallographic data processing and manuscript preparation; A.L.L., E.D.B. and N.C.J.S. were responsible for experimental design and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Discussion and Supplementary Figures 1–4 (PDF 734 kb)
Rights and permissions
About this article
Cite this article
Lovering, A., Lin, LC., Sewell, E. et al. Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis. Nat Struct Mol Biol 17, 582–589 (2010). https://doi.org/10.1038/nsmb.1819
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1819
This article is cited by
-
A multi-enzyme machine polymerizes the Haemophilus influenzae type b capsule
Nature Chemical Biology (2023)
-
Steerable artificial magnetic bacteria with target delivery ability of calcium carbonate for soil improvement
Applied Microbiology and Biotechnology (2023)
-
Attachment efficiency of gold nanoparticles by Gram-positive and Gram-negative bacterial strains governed by surface charges
Journal of Nanoparticle Research (2019)