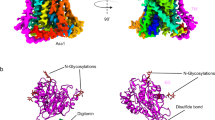
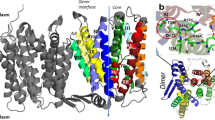
Abstract
CaiT is a membrane antiporter that catalyzes the exchange of l-carnitine with γ-butyrobetaine across the Escherichia coli membrane. To obtain structural insights into the antiport mechanism, we solved the crystal structure of CaiT at a resolution of 3.15 Å. We crystallized CaiT as a homotrimer complex, in which each protomer contained 12 transmembrane helices and 4 l-carnitine molecules outlining the transport pathway across the membrane. Mutagenesis studies revealed a primary binding site at the center of the protein and a secondary substrate-binding site at the bottom of the intracellular vestibule. These results, together with the insights obtained from structural comparison with structurally homologous transporters, provide mechanistic insights into the association between substrate translocation and the conformational changes of CaiT.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Poolman, B. Precursor/product antiport in bacteria. Mol. Microbiol. 4, 1629–1636 (1990).
Law, C.J., Maloney, P.C. & Wang, D.N. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62, 289–305 (2008).
Dierks, T., Riemer, E. & Krämer, R. Reaction mechanism of the reconstituted aspartate/glutamate carrier from bovine heart mitochondria. Biochim. Biophys. Acta 943, 231–244 (1988).
Anantharam, V., Allison, M.J. & Maloney, P.C. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J. Biol. Chem. 264, 7244–7250 (1989).
Poolman, B., Royer, T.J., Mainzer, S.E. & Schmidt, B.F. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J. Bacteriol. 171, 244–253 (1989).
Pebay-Peyroula, E. et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 (2003).
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966).
Jencks, W.P. The utilization of binding energy in coupled vectorial processes. Adv. Enzymol. 51, 75–106 (1980).
Tanford, C. Mechanism of free energy coupling in active transport. Annu. Rev. Biochem. 52, 379–409 (1983).
Fritz, I.B. & Yue, K.T. Effects of carnitine on acetyl-coA oxidation by heart muscle mitochondria. Am. J. Physiol. 206, 531–535 (1964).
Treem, W.R., Stanley, C.A., Finegold, D.N., Hale, D.E. & Coates, P.M. Primary carnitine deficiency due to a failure of carnitine transport in kidney, muscle, and fibroblasts. N. Engl. J. Med. 319, 1331–1336 (1988).
Nalecz, K.A., Miecz, D., Berezowski, V. & Cecchelli, R. Carnitine: transport and physiological functions in the brain. Mol. Aspects Med. 25, 551–567 (2004).
Jung, K., Jung, H. & Kleber, H.P. Regulation of l-carnitine metabolism in Escherichia coli. J. Basic Microbiol. 27, 131–137 (1987).
Kleber, H.P. Bacterial carnitine metabolism. FEMS Microbiol. Lett. 147, 1–9 (1997).
Seim, H., Ezold, R., Kleber, H.P. & Strack, E. Metabolism of l-carnitine in enterobacteria. Z. Allg. Mikrobiol. 20, 591–594 (1980).
Tamai, I. et al. Cloning and characterization of a novel human pH-dependent organic cation transporter, OCTN1. FEBS Lett. 419, 107–111 (1997).
Wu, X., Prasad, P.D., Leibach, F.H. & Ganapathy, V. cDNA sequence, transport function, and genomic organization of human OCTN2, a new member of the organic cation transporter family. Biochem. Biophys. Res. Commun. 246, 589–595 (1998).
Tamai, I. et al. Molecular and functional identification of sodium ion-dependent, high affinity human carnitine transporter OCTN2. J. Biol. Chem. 273, 20378–20382 (1998).
Tein, I. Carnitine transport: pathophysiology and metabolism of known molecular defects. J. Inherit. Metab. Dis. 26, 147–169 (2003).
Jung, H. et al. CaiT of Escherichia coli, a new transporter catalyzing l-carnitine/γ-butyrobetaine exchange. J. Biol. Chem. 277, 39251–39258 (2002).
Peter, H., Burkovski, A. & Krämer, R. Isolation, characterization, and expression of the Corynebacterium glutamicum betP gene, encoding the transport system for the compatible solute glycine betaine. J. Bacteriol. 178, 5229–5234 (1996).
Kappes, R.M., Kempf, B. & Bremer, E. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178, 5071–5079 (1996).
Boscari, A., Mandon, K., Dupont, L., Poggi, M.C. & Le Rudulier, D. BetS is a major glycine betaine/proline betaine transporter required for early osmotic adjustment in Sinorhizobium meliloti. J. Bacteriol. 184, 2654–2663 (2002).
Vinothkumar, K.R., Raunser, S., Jung, H. & Kühlbrandt, W. Oligomeric structure of the carnitine transporter CaiT from Escherichia coli. J. Biol. Chem. 281, 4795–4801 (2006).
Ressl, S. et al. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature 458, 47–52 (2009).
Yamashita, A. et al. Crystal structure of a bacterial homologue of Na+/Cl−dependent neurotransmitter transporters. Nature 437, 215–223 (2005).
Faham, S. et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321, 810–814 (2008).
Weyand, S. et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322, 709–713 (2008).
Fang, Y. et al. Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature 460, 1040–1043 (2009).
Gao, X. et al. Structure and mechanism of an amino acid antiporter. Science 324, 1565–1568 (2009).
Shaffer, P.L., Goehring, A., Shankaranarayanan, A. & Gouaux, E. Structure and mechanism of a Na+-independent amino acid transporter. Science 325, 1010–1014 (2009).
Schiefner, A. et al. Cation-p interactions as determinants for binding of the compatible solutes betaine and proline betaine by the periplasmic ligand-binding protein ProX from Escherichia coli. J. Biol. Chem. 279, 5588–5596 (2004).
Singh, S.K., Piscitelli, C.L., Yamashita, A. & Gouaux, E.A. Competitive inhibitor traps LeuT in an open-to-out conformation. Science 322, 1655–1661 (2008).
Shi, L., Quick, M., Zhao, Y., Weinstein, H. & Javitch, J.A. The mechanism of a neurotransmitter: sodium symporter-inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell 30, 667–677 (2008).
Quick, M. et al. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc. Natl. Acad. Sci. USA 106, 5563–5568 (2009).
DeLano, W.L. The PyMOL Molecular Graphics System. (DeLano Scientific, San Carlos, California, USA, 2002) <http://www.pymol.org.>.
Drew, D., Lerch, M., Kunji, E., Slotboom, D.J. & de Gier, J.W. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods 3, 303–313 (2006).
Collaborative Computational Project. Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007).
Terwilliger, T.C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008).
Cowtan, K. An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography, DM 31, 34–38 (1994).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Jung, H., Tebbe, S., Schmid, R. & Jung, K. Unidirectional reconstitution and characterization of purified Na+/proline transporter of Escherichia coli. Biochemistry 37, 11083–11088 (1998).
Acknowledgements
We thank the staff at the European Synchrotron Radiation Facility in France, the Spring-8 (beamline BL41XU) in Japan and the Swiss Light Source (beamline X06SA) in Switzerland for help with data collection, J. Tang and Y.-B. Wang for technical support and J. Chen for discussions. This research was financially supported by the National Key Basic Research Program (grant numbers 2009CB918600, 2009CB918803), the National Natural Science Foundation of China (grant number 30721003), the National High Technology Research and Development Program of China (grant number 2006AA02A319) and the Knowledge Innovation Program of the Chinese Academy of Sciences (grant number KSCX2-YW-R-123).
Author information
Authors and Affiliations
Contributions
L.T. performed expression, purification, crystallization and the crystal structure determination; L.B. and W.-h.W. contributed to vector construction and protein expression; L.B. performed the mutagenesis studies and transport activity measurements; T.J. and L.T. designed the research; T.J. supervised the work; L.T. and T.J. analyzed the results and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 612 kb)
Rights and permissions
About this article
Cite this article
Tang, L., Bai, L., Wang, Wh. et al. Crystal structure of the carnitine transporter and insights into the antiport mechanism. Nat Struct Mol Biol 17, 492–496 (2010). https://doi.org/10.1038/nsmb.1788
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1788