Abstract
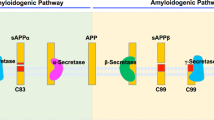
γ-secretase is an aspartyl protease that cleaves multiple substrates within their transmembrane domains. γ-secretase processes the amyloid precursor protein (APP) to generate γ-amyloid (Aγ) peptides associated with Alzheimer's disease. Here, we show that APP possesses a substrate inhibitory domain (ASID) that negatively modulates γ-secretase activity for Aγ production by binding to an allosteric site within the γ-secretase complex. Alteration of this ASID by deletion or mutation, as is seen with the Flemish mutation (A21G), reduces its inhibitory potency and promotes Aγ production. Notably, peptides derived from ASID show selective inhibition of γ-secretase activity for Aγ production over Notch1 processing. Therefore, this mode of regulation represents an unprecedented mechanism for modulating γ-secretase, providing insight into the molecular basis of Alzheimer's disease pathogenesis and a potential strategy for the development of therapeutics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hardy, J. & Allsop, D. Amyloid deposition as the central event in the etiology of Alzheimer's disease. Trends Pharmacol. Sci. 12, 383–388 (1991).
Jarrett, J.T., Berger, E.P. & Lansbury, P.T., Jr. The C-terminus of the β protein is critical in amyloidogenesis. Ann. NY Acad. Sci. 695, 144–148 (1993).
Zhao, G. et al. Identification of a new presenilin-dependent ζ-cleavage site within the transmembrane domain of amyloid precursor protein. J. Biol. Chem. 279, 50647–50650 (2004).
Sastre, M. et al. Presenilin-dependent γ-secretase processing of β-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2, 835–841 (2001).
Cao, X. & Sudhof, T.C. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293, 115–120 (2001).
De Strooper, B. Aph-1, Pen-2, and nicastrin with presenilin generate an active γ-secretase complex. Neuron 38, 9–12 (2003).
Li, Y.M. et al. Photoactivated γ-secretase inhibitors directed to the active site covalently label presenilin 1. Nature 405, 689–694 (2000).
Wolfe, M.S. et al. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature 398, 513–517 (1999).
Esler, W.P. et al. Transition-state analogue inhibitors of γ-secretase bind directly to presenilin-1. Nat. Cell Biol. 2, 428–434 (2000).
Thinakaran, G. et al. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron 17, 181–190 (1996).
Brown, M.S., Ye, J., Rawson, R.B. & Goldstein, J.L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–398 (2000).
Struhl, G. & Adachi, A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol. Cell 6, 625–636 (2000).
Shah, S. et al. Nicastrin functions as a γ-secretase-substrate receptor. Cell 122, 435–447 (2005).
Selkoe, D.J. Alzheimer's disease: genes, proteins, and therapy. Physiol. Rev. 81, 741–766 (2001).
Kowalska, A. Amyloid precursor protein gene mutations responsible for early-onset autosomal dominant Alzheimer's disease. Folia Neuropathol. 41, 35–40 (2003).
Goate, A. et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature 349, 704–706 (1991).
Vassar, R. et al. β-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741 (1999).
Suzuki, N. et al. An increased percentage of long amyloid β protein secreted by familial amyloid β protein precursor (β APP717) mutants. Science 264, 1336–1340 (1994).
De Jonghe, C. et al. Pathogenic APP mutations near the γ-secretase cleavage site differentially affect Aβ secretion and APP C-terminal fragment stability. Hum. Mol. Genet. 10, 1665–1671 (2001).
Lichtenthaler, S.F., Ida, N., Multhaup, G., Masters, C.L. & Beyreuther, K. Mutations in the transmembrane domain of APP altering γ-secretase specificity. Biochemistry 36, 15396–15403 (1997).
Maruyama, K. et al. Familial Alzheimer's disease–linked mutations at Val717 of amyloid precursor protein are specific for the increased secretion of Aβ42(43). Biochem. Biophys. Res. Commun. 227, 730–735 (1996).
Jankowsky, J.L. et al. Mutant presenilins specifically elevate the levels of the 42 residue β-amyloid peptide in vivo: evidence for augmentation of a 42-specific γ-secretase. Hum. Mol. Genet. 13, 159–170 (2004).
Yin, Y.I. et al. γ-Secretase substrate concentration modulates the Aβ42/Aβ40 ratio: implications for Alzeheimer's disease. J. Biol. Chem. 282, 23639–23644 (2007).
Haass, C., Hung, A.Y., Selkoe, D.J. & Teplow, D.B. Mutations associated with a locus for familial Alzheimer's disease result in alternative processing of amyloid β–protein precursor. J. Biol. Chem. 269, 17741–17748 (1994).
De Jonghe, C. et al. Flemish and Dutch mutations in amyloid β precursor protein have different effects on amyloid β secretion. Neurobiol. Dis. 5, 281–286 (1998).
Nilsberth, C. et al. The 'Arctic' APP mutation (E693G) causes Alzheimer's disease by enhanced Aβ protofibril formation. Nat. Neurosci. 4, 887–893 (2001).
Tian, G. et al. Linear non-competitive inhibition of solubilized human γ-secretase by pepstatin A methylester, L685458, sulfonamides, and benzodiazepines. J. Biol. Chem. 277, 31499–31505 (2002).
Kornilova, A.Y., Bihel, F., Das, C. & Wolfe, M.S. The initial substrate-binding site of γ-secretase is located on presenilin near the active site. Proc. Natl. Acad. Sci. USA 102, 3230–3235 (2005).
Li, Y.M. et al. Presenilin 1 is linked with γ-secretase activity in the detergent solubilized state. Proc. Natl. Acad. Sci. USA 97, 6138–6143 (2000).
Brody, D.L. et al. Amyloid-β dynamics correlate with neurological status in the injured human brain. Science 321, 1221–1224 (2008).
Lichtenthaler, S.F., Multhaup, G., Masters, C.L. & Beyreuther, K. A novel substrate for analyzing Alzheimer's disease γ-secretase. FEBS Lett. 453, 288–292 (1999).
Segel, I.H. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems 22nd edn. (Wiley, New York, 1993).
Li, T., Ma, G., Cai, H., Price, D.L. & Wong, P.C. Nicastrin is required for assembly of presenilin/γ-secretase complexes to mediate Notch signaling and for processing and trafficking of β-amyloid precursor protein in mammals. J. Neurosci. 23, 3272–3277 (2003).
Donoviel, D.B. et al. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13, 2801–2810 (1999).
Lai, M.T. et al. Presenilin-1 and presenilin-2 exhibit distinct yet overlapping γ-secretase activities. J. Biol. Chem. 278, 22475–22481 (2003).
Dorman, G. & Prestwich, G.D. Benzophenone photophores in biochemistry. Biochemistry 33, 5661–5673 (1994).
Chun, J., Yin, Y.I., Yang, G., Tarassishin, L. & Li, Y.M. Stereoselective synthesis of photoreactive peptidomimetic γ-secretase inhibitors. J. Org. Chem. 69, 7344–7347 (2004).
Yang, G. et al. Stereo-controlled synthesis of novel photoreactive γ-secretase inhibitors. Bioorg. Med. Chem. Lett. 19, 922–925 (2009).
Das, C. et al. Designed helical peptides inhibit an intramembrane protease. J. Am. Chem. Soc. 125, 11794–11795 (2003).
Kukar, T.L. et al. Substrate-targeting γ-secretase modulators. Nature 453, 925–929 (2008).
Soto, C., Kindy, M.S., Baumann, M. & Frangione, B. Inhibition of Alzheimer's amyloidosis by peptides that prevent β-sheet conformation. Biochem. Biophys. Res. Commun. 226, 672–680 (1996).
Chorev, M. The partial retro-inverso modification: a road traveled together. Biopolymers 80, 67–84 (2005).
Guichard, G. et al. Antigenic mimicry of natural L-peptides with retro-inverso-peptidomimetics. Proc. Natl. Acad. Sci. USA 91, 9765–9769 (1994).
Lee, Y.S. et al. Partial retro-inverso, retro, and inverso modifications of hydrazide linked bifunctional peptides for opioid and cholecystokinin (CCK) receptors. J. Med. Chem. 50, 165–168 (2007).
Sakurai, K., Chung, H.S. & Kahne, D. Use of a retroinverso p53 peptide as an inhibitor of MDM2. J. Am. Chem. Soc. 126, 16288–16289 (2004).
Schroeter, E.H., Kisslinger, J.A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998).
Krishnaswamy, S. Exosite-driven substrate specificity and function in coagulation. J. Thromb. Haemost. 3, 54–67 (2005).
Overall, C.M. Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol. Biotechnol. 22, 51–86 (2002).
Mayer, S.C. et al. Discovery of begacestat, a Notch-1-sparing γ-secretase inhibitor for the treatment of Alzheimer's disease. J. Med. Chem. 51, 7348–7351 (2008).
Cole, D.C. et al. (S)-N-(5-chlorothiophene-2-sulfonyl)-β,β-diethylalaninol a Notch-1-sparing γ-secretase inhibitor. Bioorg. Med. Chem. Lett. 19, 926–929 (2009).
Bhattacharyya, R.P., Remenyi, A., Yeh, B.J. & Lim, W.A. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 75, 655–680 (2006).
Dickens, M. et al. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science 277, 693–696 (1997).
Kaneto, H. et al. Possible novel therapy for diabetes with cell-permeable JNK-inhibitory peptide. Nat. Med. 10, 1128–1132 (2004).
Balendran, A. et al. A 3-phosphoinositide-dependent protein kinase-1 (PDK1) docking site is required for the phosphorylation of protein kinase Czeta (PKCzeta) and PKC-related kinase 2 by PDK1. J. Biol. Chem. 275, 20806–20813 (2000).
Balendran, A. et al. PDK1 acquires PDK2 activity in the presence of a synthetic peptide derived from the carboxyl terminus of PRK2. Curr. Biol. 9, 393–404 (1999).
Kaidanovich-Beilin, O. & Eldar-Finkelman, H. Peptides targeting protein kinases: strategies and implications. Physiology (Bethesda) 21, 411–418 (2006).
Pufall, M.A. & Graves, B.J. Autoinhibitory domains: modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 18, 421–462 (2002).
McCafferty, D.G., Lessard, I.A. & Walsh, C.T. Mutational analysis of potential zinc-binding residues in the active site of the enterococcal D-Ala-D-Ala dipeptidase VanX. Biochemistry 36, 10498–10505 (1997).
Hecimovic, S. et al. Mutations in APP have independent effects on Aβ and CTFγ generation. Neurobiol. Dis. 17, 205–218 (2004).
Acknowledgements
We thank D. Scheinberg, J. Shafer, R. Zhou and S. Gardell for advice and comments on the manuscript, C.C. Shelton, L. Placanica and C. Crump for discussions and suggestions regarding this manuscript, H. Zheng (Baylor College of Medicine) for the APPc antibody, M.-t. Lai (Merck Research Laboratories) for the PS-1 NTF antibody, C.T. Walsh (Harvard Medical School) for the pIAD16 plasmid and R. Kopan for the ΔE-Notch1 construct (Washington University School of Medicine). This work is supported by US National Institutes of Health grant AG026660 (Y.-M.L.), the Alzheimer's Association (Zenith Fellows Award to Y.-M.L.) and the American Health Assistance Foundation (Y.-M.L.), the Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center and the William Randolph Hearst Fund in Experimental Therapeutics.
Author information
Authors and Affiliations
Contributions
Y.T. and Y.-M.L designed the experiments and prepared this manuscript; Y.T. and D.C. synthesized and characterized the peptides; Y.T and B.B. made the constructs, purified the proteins and performed in vitro and cell-based assays; Y.T. conducted the kinetic and photoaffinity labeling experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 334 kb)
Rights and permissions
About this article
Cite this article
Tian, Y., Bassit, B., Chau, D. et al. An APP inhibitory domain containing the Flemish mutation residue modulates γ-secretase activity for Aβ production. Nat Struct Mol Biol 17, 151–158 (2010). https://doi.org/10.1038/nsmb.1743
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1743
This article is cited by
-
Genetic Phenotypes of Alzheimer’s Disease: Mechanisms and Potential Therapy
Phenomics (2023)
-
APP-BACE1 Interaction and Intracellular Localization Regulate Aβ Production in iPSC-Derived Cortical Neurons
Cellular and Molecular Neurobiology (2023)
-
Hypoxia Inducible Factor-1α binds and activates γ-secretase for Aβ production under hypoxia and cerebral hypoperfusion
Molecular Psychiatry (2022)
-
Clusterin transduces Alzheimer-risk signals to amyloidogenesis
Signal Transduction and Targeted Therapy (2022)
-
The innate immunity protein IFITM3 modulates γ-secretase in Alzheimer’s disease
Nature (2020)