Abstract
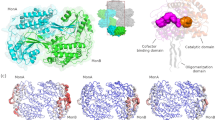
In approximately one billion people, a point mutation inactivates a key detoxifying enzyme, aldehyde dehydrogenase (ALDH2). This mitochondrial enzyme metabolizes toxic biogenic and environmental aldehydes, including the endogenously produced 4-hydroxynonenal (4HNE) and the environmental pollutant acrolein, and also bioactivates nitroglycerin. ALDH2 is best known, however, for its role in ethanol metabolism. The accumulation of acetaldehyde following the consumption of even a single alcoholic beverage leads to the Asian alcohol-induced flushing syndrome in ALDH2*2 homozygotes. The ALDH2*2 allele is semidominant, and heterozygotic individuals show a similar but less severe phenotype. We recently identified a small molecule, Alda-1, that activates wild-type ALDH2 and restores near-wild-type activity to ALDH2*2. The structures of Alda-1 bound to ALDH2 and ALDH2*2 reveal how Alda-1 activates the wild-type enzyme and how it restores the activity of ALDH2*2 by acting as a structural chaperone.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hurley, T.D., Edenberg, H.J. & Li, T.K. (2002) Pharmacogenomics of alcoholism. in Pharmacogenomics: The Search for Individualized Therapies (Licinio, J., & Wong, M., eds.) 417–441 (2002).
Chen, Z. et al. An essential role for mitochondrial aldehyde dehydrogenase in nitroglycerin bioactivation. Proc. Natl. Acad. Sci. USA 102, 12159–12164 (2005).
Chen, C.H. et al. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science 321, 1493–1495 (2008).
Higuchi, S. et al. Aldehyde dehydrogenase genotypes in Japanese alcoholics. Lancet 343, 741–742 (1994).
Brooks, P.J., Enoch, M.-A., Goldman, D., Li, T.-K. & Yokoyama, A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 6, 258–263 (2009).
Li, Y. et al. Mitochondrial aldehyde dehydrogenase-2 (ALDH2) Glu504Lys polymorphism contributes to the variation in efficacy of sublingual nitroglycerin. J. Clin. Invest. 116, 506–511 (2006).
Jo, S.A. et al. Glu487Lys polymorphism in the gene for mitochondrial aldehyde dehydrogenase 2 is associated with myocardial infarction in elderly Korean men. Clin. Chim. Acta 382, 43–47 (2007).
Marchitti, S.A., Deitrich, R.A. & Vasiliou, V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol. Rev. 59, 125–150 (2007).
Larson, H.N., Weiner, H. & Hurley, T.D. Disruption of the coenzyme binding site and dimer interface revealed in the crystal structure of mitochondrial aldehyde dehydrogenase “Asian” variant. J. Biol. Chem. 280, 30550–30556 (2005).
Farres, J. et al. Effect of changing glutamate to lysine in rat and human liver mitochondrial aldehyde dehydrogenase: a model to study human (Oriental type) class 2 aldehyde dehydrogenase. J. Biol. Chem. 269, 13854–13860 (1994).
Larson, H. et al. Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487. J. Biol. Chem. 282, 12940–12950 (2007).
Suzuki, Y., Ogawa, S. & Sakakibara, Y. Chaperone therapy for neuronopathic lysosomal diseases: competitive inhibitors as chemical chaperones for enchancement of mutant enzyme activities. Perspect. Medicin. Chem. 3, 7–19 (2009).
Lowe, E.D., Gao, G.-Y., Johnson, L.N. & Keung, W.M. Structure of daidzin, a naturally occurring anti–alcohol-addiction agent, in complex with human mitochondrial aldehyde dehydrogenase. J. Med. Chem. 51, 4482–4487 (2008).
Hammen, P.K., Allali-Hassani, A., Hallenga, K., Hurley, T.D. & Weiner, H. Multiple conformations of NAD and NADH when bound to human cytosolic and mitochondrial aldehyde dehydrogenase. Biochemistry 41, 7156–7168 (2002).
Perez-Miller, S. & Hurley, T.D. Coenzyme isomerization is integral to catalysis in human aldehyde dehydrogenase. Biochemistry 42, 7100–7109 (2003).
Liu, Z.-J. et al. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 4, 317–326 (1997).
Lamb, A.L. & Newcomer, M.E. The structure of retinal dehydrogenase type II at 2.7 Å resolution: implications for retinal specificity. Biochemistry 38, 6003–6011 (1999).
D'Ambrosio, K. et al. The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis. Biochemistry 45, 2978–2986 (2006).
Feldman, R.I. & Weiner, H. Horse liver aldehyde dehydrogenase. II. Kinetics and mechanistic implications of the dehydrogenase and esterase activity. J. Biol. Chem. 247, 267–272 (1972).
Takahashi, K. & Weiner, W. Nicotinamide adenine dinucleotide activation of the esterase reaction of horse liver aldehyde dehydrogenase. Biochemistry 20, 2720–2726 (1981).
Wenzel, P. et al. ALDH-2 deficiency increases cardiovascular oxidative stress–evidence for indirect antioxidative properties. Biochem. Biophys. Res. Commun. 367, 137–143 (2008).
Jinsmaa, Y. et al. Products of oxidative stress inhibit aldehyde oxidation and reduction pathways in dopamine catabolism yielding elevated levels of a reactive intermediate. Chem. Res. Toxicol. 22, 835–841 (2009).
Marchal, S. & Branlant, G. Evidence for the chemical activation of essential Cys302 upon cofactor binding to nonphosphorylating glyceraldehyde 3-phosphate dehydrogenase from Streptococcus mutans. Biochemistry 38, 12950–12958 (1999).
Mukerjee, N. & Pietruszko, R. Human mitochondrial aldehyde dehydrogenase substrate specificity: comparison of esterase with dehydrogenase reaction. Arch. Biochem. Biophys. 299, 23–29 (1992).
DeLano, W.L. The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, California, USA, 2002).
Ni, L., Zhou, J., Hurley, T.D. & Weiner, H. Human liver mitochondrial aldehyde dehydrogenase: three-dimensional structure and the restoration of solubility and activity of chimeric forms. Protein Sci. 8, 2784–2790 (1999).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Acknowledgements
T.D.H. wishes to thank the staff at the 19-ID beamline facility, especially M. Cuff, N. Duke and S. Ginell. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the US Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357. This work was supported in part by US National Institutes of Health grants AA011982 and AA018123 to T.D.H. and AA11147 to D.M.-R.; S.P.-M. was supported by US National Institutes of Health training fellowship T32-DK064466.
Author information
Authors and Affiliations
Contributions
S.P.-M. performed experiments, data analysis and manuscript preparation; H.Y. performed experiments and helped with data analysis; R.V. performed experiments; C.-H.C. helped with data analysis and manuscript preparation; D.M.-R. initiated the study and helped with study design, data analysis and manuscript preparation; T.D.H. designed the study and performed data analysis, experiments and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
C.-H.C. and D.M.-R. plan to found a pharmaceutical company based on the activators of aldehyde dehydrogenases. Neither author holds any financial interest at the moment.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 (PDF 38 kb)
Rights and permissions
About this article
Cite this article
Perez-Miller, S., Younus, H., Vanam, R. et al. Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat Struct Mol Biol 17, 159–164 (2010). https://doi.org/10.1038/nsmb.1737
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1737
This article is cited by
-
A common East-Asian ALDH2 mutation causes metabolic disorders and the therapeutic effect of ALDH2 activators
Nature Communications (2023)
-
Impaired dopamine metabolism in Parkinson’s disease pathogenesis
Molecular Neurodegeneration (2019)
-
Effects of the common polymorphism in the human aldehyde dehydrogenase 2 (ALDH2) gene on the lung
Respiratory Research (2017)
-
Aldehyde dehydrogenase 2 activation and coevolution of its εPKC-mediated phosphorylation sites
Journal of Biomedical Science (2017)
-
ALDH2 polymorphism and alcohol-related cancers in Asians: a public health perspective
Journal of Biomedical Science (2017)