Abstract
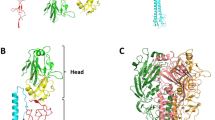
The highly contagious measles virus infects millions of individuals worldwide, causing serious disease in children of developing countries. Infection is initiated by attachment of the measles virus hemagglutinin (MV-H), a glycoprotein anchored to the virus envelope, to the host cell receptors CD46 or signaling lymphocyte activation molecule (SLAM). Here we report the crystal structure of MV-H in complex with a CD46 protein spanning the two N-terminal domains. A unique groove at the side of the MV-H β-propeller domain, which is absent in homologous paramyxovirus attachment proteins, engages residues in both CD46 domains. Key contacts involve a protruding loop in the N-terminal CD46 domain that carries two sequential proline residues (PP motif) and penetrates deeply into a hydrophobic socket in MV-H. We identify a similar PP motif in SLAM, defining a common measles virus recognition epitope in the CD46 and SLAM receptor proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Griffin, D.E. Measles virus. in Fields Virology Vol. 1 (eds. Fields, B.N. et al.) 1551–1585 (Lippincott, Williams & Wilkins, Philadelphia, 2007).
Rota, P.A., Featherstone, D.A. & Bellini, W.J. Molecular epidemiology of measles virus. Curr. Top. Microbiol. Immunol. 330, 129–150 (2009).
Naniche, D. et al. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 67, 6025–6032 (1993).
Dorig, R.E., Marcil, A., Chopra, A. & Richardson, C.D. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75, 295–305 (1993).
Manchester, M. et al. Measles virus recognizes its receptor, CD46, via two distinct binding domains within SCR1–2. Virology 232, 1–12 (1997).
Buchholz, C.J. et al. Mapping of the primary binding site of measles virus to its receptor CD46. J. Biol. Chem. 272, 22072–22079 (1997).
Casasnovas, J.M., Larvie, M. & Stehle, T. Crystal structure of two CD46 domains reveals an extended measles virus-binding surface. EMBO J. 18, 2911–2922 (1999).
Tatsuo, H., Ono, N., Tanaka, K. & Yanagi, Y. SLAM (CDw 150) is a cellular receptor for measles virus. Nature 406, 893–897 (2000).
Ono, N., Tatsuo, H., Tanaka, K., Minagawa, H. & Yanagi, Y. V domain of human SLAM (CDw150) is essential for its function as a measles virus receptor. J. Virol. 75, 1594–1600 (2001).
Manchester, M. et al. Clinical isolates of measles virus use CD46 as a cellular receptor. J. Virol. 74, 3967–3974 (2000).
Erlenhofer, C., Duprex, W.P., Rima, B.K., ter Meulen, V. & Schneider-Schaulies, J. Analysis of receptor (CD46, CD150) usage by measles virus. J. Gen. Virol. 83, 1431–1436 (2002).
Lecouturier, V. et al. Identification of two amino acids in the hemagglutinin glycoprotein of measles virus (MV) that govern hemadsorption, HeLa cell fusion, and CD46 downregulation: phenotypic markers that differentiate vaccine and wild-type MV strains. J. Virol. 70, 4200–4204 (1996).
Shibahara, K., Hotta, H., Katayama, Y. & Homma, M. Increased binding activity of measles virus to monkey red blood cells after long-term passage in Vero cell cultures. J. Gen. Virol. 75, 3511–3516 (1994).
Tahara, M., Takeda, M., Seki, F., Hashiguchi, T. & Yanagi, Y. Multiple amino acid substitutions in hemagglutinin are necessary for wild-type measles virus to acquire the ability to use receptor CD46 efficiently. J. Virol. 81, 2564–2572 (2007).
Rota, J.S., Wang, Z.D., Rota, P.A. & Bellini, W.J. Comparison of sequences of the H, F, and N coding genes of measles virus vaccine strains. Virus Res. 31, 317–330 (1994).
Schneider, U., von Messling, V., Devaux, P. & Cattaneo, R. Efficiency of measles virus entry and dissemination through different receptors. J. Virol. 76, 7460–7467 (2002).
Santiago, C., Björling, E., Stehle, T. & Casasnovas, J.M. Distinct kinetics for binding of the CD46 and SLAM receptors to overlapping sites in the measles virus hemagglutinin protein. J. Biol. Chem. 277, 32294–32301 (2002).
Hashiguchi, T. et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. USA 104, 19535–19540 (2007).
Masse, N. et al. Measles virus (MV) hemagglutinin: evidence that attachment sites for MV receptors SLAM and CD46 overlap on the globular head. J. Virol. 78, 9051–9063 (2004).
Vongpunsawad, S., Oezgun, N., Braun, W. & Cattaneo, R. Selectively receptor-blind measles viruses: identification of residues necessary for SLAM- or CD46-induced fusion and their localization on a new hemagglutinin structural model. J. Virol. 78, 302–313 (2004).
Colf, L.A., Juo, Z.S. & Garcia, K.C. Structure of the measles virus hemagglutinin. Nat. Struct. Mol. Biol. 14, 1227–1228 (2007).
Leonard, V.H.J. et al. Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J. Clin. Invest. 118, 2448–2458 (2008).
Tahara, M. et al. Measles virus infects both polarized epithelial and immune cells by using distinctive receptor-binding sites on its hemagglutinin. J. Virol. 82, 4630–4637 (2008).
Persson, B.D. et al. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat. Struct. Mol. Biol. 14, 164–166 (2007).
Maisner, A. et al. The N-glycan of the SCR 2 region is essential for membrane cofactor protein (CD46) to function as a measles virus receptor. J. Virol. 70, 4973–4977 (1996).
Crennell, S., Takimoto, T., Portner, A. & Taylor, G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7, 1068–1074 (2000).
Bowden, T.A. et al. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 15, 567–572 (2008).
Xu, K. et al. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Acad. Sci. USA 105, 9953–9958 (2008).
Stehle, T. & Casasnovas, J.M. Specificity switching in virus-receptor complexes. Curr. Opin. Struct. Biol. 19, 181–188 (2009).
Ono, N., Tatsuo, H., Tanaka, K., Minagawa, H. & Yanagi, Y. V domain of human SLAM (CDw150) is essential for its function as a measles virus receptor. J. Virol. 75, 1594–1600 (2001).
Kwong, P.D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998).
Wang, J. et al. Atomic structure of a fragment of human CD4 containing two immunoglobulin-like domains. Nature 348, 411–418 (1990).
Bergelson, J.M. et al. Coxsackievirus B3 adapted to growth in RD cells binds to decay-accelerating factor (CD55). J. Virol. 69, 1903–1906 (1995).
Hueffer, K. & Parrish, C.R. Parvovirus host range, cell tropism and evolution. Curr. Opin. Microbiol. 6, 392–398 (2003).
Martinez, M.A., Verdaguer, N., Mateu, M.G. & Domingo, E. Evolution subverting essentiality: dispensability of the cell attachment Arg-Gly-Asp motif in multiply passaged foot-and-mouth disease virus. Proc. Natl. Acad. Sci. USA 94, 6798–6802 (1997).
Manchester, M., Naniche, D. & Stehle, T. CD46 as measles receptor: form follows function. Virology 274, 5–10 (2000).
Lawrence, M.C. et al. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 335, 1343–1357 (2004).
Yuan, P. et al. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13, 803–815 (2005).
Xiong, J.P. et al. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science 294, 339–345 (2001).
Sondek, J., Bohm, A., Lambright, D.G., Hamm, H.E. & Sigler, P.B. Crystal structure of a G-protein β γ dimer at 2.1A resolution. Nature 379, 369–374 (1996).
Ertl, O.T., Wenz, D.C., Bouche, F.B., Berbers, G.A. & Muller, C.P. Immunodominant domains of the measles virus hemagglutinin protein eliciting a neutralizing human B cell response. Arch. Virol. 148, 2195–2206 (2003).
Hu, A., Sheshberadaran, H., Norrby, E. & Kövamees, J. Molecular characterization of epitopes on the measles virus hemagglutinin protein. Virology 192, 351–354 (1993).
Hummel, K.B. & Bellini, W.J. Localization of monoclonal antibody epitopes and functional domains in the hemagglutinin protein of measles virus. J. Virol. 69, 1913–1916 (1995).
Liebert, U.G. et al. Antigenic determinants of measles virus hemagglutinin associated with neurovirulence. J. Virol. 68, 1486–1493 (1994).
Ziegler, D. et al. Protection against measles virus encephalitis by monoclonal antibodies binding to a cystine loop domain of the H protein mimicked by peptides which are not recognized by maternal antibodies. J. Gen. Virol. 77, 2479–2489 (1996).
Stern, L.B., Greenberg, M., Gershoni, J.M. & Rozenblatt, S. The hemagglutinin envelope protein of canine distemper virus (CDV) confers cell tropism as illustrated by CDV and measles virus complementation analysis. J. Virol. 69, 1661–1668 (1995).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Read, R.J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57, 1373–1382 (2001).
Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Cao, E. et al. NTB-A receptor crystal structure: insights into homophilic interactions in the signaling lymphocytic activation molecule receptor family. Immunity 25, 559–570 (2006).
Yan, Q. et al. Structure of CD84 provides insight into SLAM family function. Proc. Natl. Acad. Sci. USA 104, 10583–10588 (2007).
van Raaij, M.J., Chouin, E., van der Zandt, H., Bergelson, J.M. & Cusack, S. Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 Å resolution. Structure 8, 1147–1155 (2000).
Acknowledgements
We are grateful to F. Pazos for assistance with secondary structure prediction and to R. Fernandez-Muñoz for helpful discussions. We acknowledge the European Molecular Biology Laboratory, the Deutsches Elektronen Synchrotron and the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities. This work has been supported by grants from the Ministerio de Ciencia e Innovación (BFU2005-05972 and BFU2008-00971) to J.M.C. T.S. acknowledges support from SFB-685.
Author information
Authors and Affiliations
Contributions
C.S. and J.M.C. designed the constructs. C.S. prepared the proteins and crystallized the MV-H–CD46 complex. C.S. and J.M.C. contributed to data collection and structure determination. C.S., T.S. and J.M.C. performed structure refinement and model building. C.S., M.L.C., T.S. and J.M.C. contributed to analysis of the data and preparation of the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Table 1 and Supplementary References (PDF 1006 kb)
Rights and permissions
About this article
Cite this article
Santiago, C., Celma, M., Stehle, T. et al. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol 17, 124–129 (2010). https://doi.org/10.1038/nsmb.1726
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1726
This article is cited by
-
Immunologic and Genetic Contributors to CD46-Dependent Immune Dysregulation
Journal of Clinical Immunology (2023)
-
Promotion of virus assembly and organization by the measles virus matrix protein
Nature Communications (2018)
-
Genome-wide associations of CD46 and IFI44L genetic variants with neutralizing antibody response to measles vaccine
Human Genetics (2017)
-
Paramyxovirus Glycoproteins and the Membrane Fusion Process
Current Clinical Microbiology Reports (2016)
-
Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4
Nature Structural & Molecular Biology (2013)