Abstract
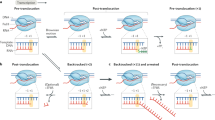
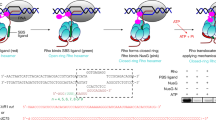
The bacterial Rho factor is a ring-shaped ATP-dependent helicase that tracks along RNA transcripts and disrupts RNA-DNA duplexes and transcription complexes in its path. Using combinatorial nucleotide analog interference mapping (NAIM), we explore the topology and dynamics of functional Rho–RNA complexes and reveal the RNA-dependent stepping mechanism of Rho helicase. Periodic Gaussian distributions of NAIM signals show that Rho forms uneven productive interactions with the track nucleotides and disrupts RNA-DNA duplexes in a succession of large (∼7-nucleotide-long) discrete steps triggered by 2′-hydroxyl activation events. This periodic 2′-OH–dependent activation does not depend on the RNA-DNA pairing energy but is finely tuned by sequence-dependent interactions with the RNA track. These features explain the strict RNA specificity and contextual efficiency of the enzyme and provide a new paradigm for conditional tracking by a helicase ring.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Pyle, A.M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 37, 317–336 (2008).
Patel, S.S. & Picha, K.M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69, 651–697 (2000).
Delagoutte, E. & von Hippel, P.H. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: integration of helicases into cellular processes. Q. Rev. Biophys. 36, 1–69 (2003).
Singleton, M.R., Dillingham, M.S. & Wigley, D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50 (2007).
Enemark, E.J. & Joshua-Tor, L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442, 270–275 (2006).
Johnson, D.S., Bai, L., Smith, B.Y., Patel, S.S. & Wang, M.D. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell 129, 1299–1309 (2007).
Galletto, R., Jezewska, M.J. & Bujalowski, W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J. Mol. Biol. 343, 83–99 (2004).
Lionnet, T., Spiering, M.M., Benkovic, S.J., Bensimon, D. & Croquette, V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. USA 104, 19790–19795 (2007).
Donmez, I. & Patel, S.S. Coupling of DNA unwinding to nucleotide hydrolysis in a ring-shaped helicase. EMBO J. 27, 1718–1726 (2008).
Richardson, J.P. Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta 1577, 251–260 (2002).
Cardinale, C.J. et al. Termination factor rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320, 935–938 (2008).
Gowrishankar, J. & Harinarayanan, R. Why is transcription coupled to translation in bacteria? Mol. Microbiol. 54, 598–603 (2004).
Bogden, C.E., Fass, D., Bergman, N., Nichols, M.D. & Berger, J.M. The structural basis for terminator recognition by the rho transcription termination factor. Mol. Cell 3, 487–493 (1999).
Hitchens, T.K., Zhan, Y., Richardson, L.V., Richardson, J.P. & Rule, G.S. Sequence-specific interactions in the RNA-binding domain of Escherichia coli transcription termination factor Rho. J. Biol. Chem. 281, 33697–33703 (2006).
Schwartz, A., Walmacq, C., Rahmouni, A.R. & Boudvillain, M. Noncanonical interactions in the management of RNA structural blocks by the transcription termination rho helicase. Biochemistry 46, 9366–9379 (2007).
Briercheck, D.M. et al. 1H, 15N and 13C resonance assignments and secondary structure determination of the RNA-binding domain of E. coli rho protein. J. Biomol. NMR 8, 429–444 (1996).
Xu, Y., Kohn, H. & Widger, W.R. Mutations in the rho transcription termination factor that affect RNA tracking. J. Biol. Chem. 277, 30023–30030 (2002).
Wei, R.R. & Richardson, J.P. Mutational changes of conserved residues in the Q-loop region of transcription factor rho greatly reduce secondary site RNA-binding. J. Mol. Biol. 314, 1007–1015 (2001).
Richardson, J.P. Activation of rho protein ATPase requires simultaneous interaction at two kinds of nucleic acid-binding sites. J. Biol. Chem. 257, 5760–5766 (1982).
Wei, R.R. & Richardson, J.P. Identification of an RNA-binding Site in the ATP binding domain of Escherichia coli Rho by H2O2/Fe-EDTA cleavage protection studies. J. Biol. Chem. 276, 28380–28387 (2001).
Skordalakes, E. & Berger, J.M. Structure of the rho transcription terminator. Mechanism of mRNA recognition and helicase loading. Cell 114, 135–146 (2003).
Skordalakes, E. & Berger, J.M. Structural insights into RNA-dependent ring closure and ATPase activation by the rho termination factor. Cell 127, 553–564 (2006).
Wang, Y. & von Hippel, P.H. Escherichia coli transcription termination factor rho. I. ATPase activation by oligonucleotide cofactors. J. Biol. Chem. 268, 13940–13946 (1993).
Walmacq, C., Rahmouni, A.R. & Boudvillain, M. Testing the steric exclusion model for hexameric helicases: substrate features that alter RNA-DNA unwinding by the transcription termination factor rho. Biochemistry 45, 5885–5895 (2006).
Richardson, L.V. & Richardson, J.P. Rho-dependent termination of transcription is governed primarily by the upstream rho utilization (rut) sequences of a terminator. J. Biol. Chem. 271, 21597–21603 (1996).
Fedorova, O., Boudvillain, M., Kawaoka, J. & Pyle, A.M. Nucleotide analog interference mapping and suppression: specific applications in studies of RNA tertiary structure, dynamic helicase mechanism and RNA-protein interactions. in Handbook of RNA Biochemistry Vol. 1 (eds. Hartmann, R.K., Bindereif, A., Schön, A. & Westhof, E.) 259–293 (Wiley-VCH, Weinheim, 2005).
Walmacq, C., Rahmouni, A.R. & Boudvillain, M. Influence of substrate composition on the helicase activity of transcription termination factor rho: reduced processivity of rho hexamers during unwinding of RNA-DNA hybrid regions. J. Mol. Biol. 342, 403–420 (2004).
Lucius, A.L., Maluf, N.K., Fischer, C.J. & Lohman, T.M. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 85, 2224–2239 (2003).
Strobel, S.A. A chemogenetic approach to RNA function/structure analysis. Curr. Opin. Struct. Biol. 9, 346–352 (1999).
Schwartz, A., Rahmouni, A.R. & Boudvillain, M. The functional anatomy of an intrinsic transcription terminator. EMBO J. 22, 3385–3394 (2003).
Freier, S.M. & Altmann, K.H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 25, 4429–4443 (1997).
Wang, S. & Kool, E.T. Origins of the large differences in stability of DNA and RNA helices: C-5 methyl and 2′-hydroxyl effects. Biochemistry 34, 4125–4132 (1995).
Wyatt, J.R. & Walker, G.T. Deoxynucleotide-containing oligoribonucleotide duplexes: stability and susceptibility to RNase V1 and RNase H. Nucleic Acids Res. 17, 7833–7842 (1989).
LeCuyer, K.A., Behlen, L.S. & Uhlenbeck, O.C. Mutagenesis of a stacking contact in the MS2 coat protein-RNA complex. EMBO J. 15, 6847–6853 (1996).
Loverix, S., Winquist, A., Stromberg, R. & Steyaert, J. An engineered ribonuclease preferring phosphorothioate RNA. Nat. Struct. Biol. 5, 365–368 (1998).
Brautigam, C.A. & Steitz, T.A. Structural principles for the inhibition of the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J. Mol. Biol. 277, 363–377 (1998).
Smith, J.S. & Nikonowicz, E.P. Phosphorothioate substitution can substantially alter RNA conformation. Biochemistry 39, 5642–5652 (2000).
Schwartz, A., Margeat, E., Rahmouni, A.R. & Boudvillain, M. Transcription termination factor rho can displace streptavidin from biotinylated RNA. J. Biol. Chem. 282, 31469–31476 (2007).
Moffitt, J.R. et al. Intersubunit coordination in a homomeric ring ATPase. Nature (2009).
Myong, S., Bruno, M.M., Pyle, A.M. & Ha, T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science 317, 513–516 (2007).
Kapanidis, A.N. et al. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314, 1144–1147 (2006).
Brieba, L.G. & Sousa, R. T7 promoter release mediated by DNA scrunching. EMBO J. 20, 6826–6835 (2001).
Galletto, R., Jezewska, M.J. & Bujalowski, W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: the effect of the 3′ arm and the stability of the dsDNA on the unwinding activity of the Escherichia coli DnaB helicase. J. Mol. Biol. 343, 101–114 (2004).
Donmez, I., Rajagopal, V., Jeong, Y.J. & Patel, S.S. Nucleic acid unwinding by hepatitis C virus and bacteriophage t7 helicases is sensitive to base pair stability. J. Biol. Chem. 282, 21116–21123 (2007).
Dutta, D., Chalissery, J. & Sen, R. Transcription termination factor rho prefers catalytically active elongation complexes for releasing RNA. J. Biol. Chem. 283, 20243–20251 (2008).
Park, J.S. & Roberts, J.W. Role of DNA bubble rewinding in enzymatic transcription termination. Proc. Natl. Acad. Sci. USA 103, 4870–4875 (2006).
Acknowledgements
We gratefully acknowledge J.M. Berger (Univ. California, Berkeley), J. Richardson (Indiana Univ.) and P.H. von Hippel (Univ. Oregon) for the gifts of plasmids and batches of purified wild-type Rho, F. Coste for help with molecular graphics, and T. Bizebard, E. Delagoutte, M. Nollmann and C. Royer for critical reading of the manuscript and helpful suggestions. This work was supported by grants from the Agence Nationale de la Recherche (PCV 2006) and the Conseil Régional du Centre (AO 2007).
Author information
Authors and Affiliations
Contributions
A.S. performed NAIM experiments and analyzed data; M.R. performed helicase kinetics and analyzed data; F.J. prepared WT and mutant Rho enzymes; E.M. performed QSAR and autocorrelation experiments, analyzed data and wrote the paper; A.R.R. analyzed data and wrote the paper; M.B. performed exploratory experiments, analyzed data and wrote the paper.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Methods (PDF 5448 kb)
Rights and permissions
About this article
Cite this article
Schwartz, A., Rabhi, M., Jacquinot, F. et al. A stepwise 2′-hydroxyl activation mechanism for the bacterial transcription termination factor Rho helicase. Nat Struct Mol Biol 16, 1309–1316 (2009). https://doi.org/10.1038/nsmb.1711
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1711
This article is cited by
-
Steps in the right direction
Nature (2009)