Abstract
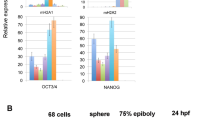
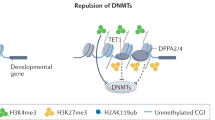
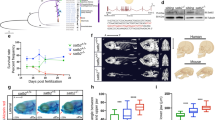
The histone variants macroH2A1 and macroH2A2 are associated with X chromosome inactivation in female mammals. However, the physiological function of macroH2A proteins on autosomes is poorly understood. Microarray-based analysis in human male pluripotent cells uncovered occupancy of both macroH2A variants at many genes encoding key regulators of development and cell fate decisions. On these genes, the presence of macroH2A1+2 is a repressive mark that overlaps locally and functionally with Polycomb repressive complex 2. We demonstrate that macroH2A1+2 contribute to the fine-tuning of temporal activation of HOXA cluster genes during neuronal differentiation. Furthermore, elimination of macroH2A2 function in zebrafish embryos produced severe but specific phenotypes. Taken together, our data demonstrate that macroH2A variants constitute an important epigenetic mark involved in the concerted regulation of gene expression programs during cellular differentiation and vertebrate development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Wolffe, A.P. & Matzke, M.A. Epigenetics: regulation through repression. Science 286, 481–486 (1999).
Sarma, K. & Reinberg, D. Histone variants meet their match. Nat. Rev. Mol. Cell Biol. 6, 139–149 (2005).
Chakravarthy, S. et al. Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 25, 7616–7624 (2005).
Costanzi, C. & Pehrson, J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393, 599–601 (1998).
Angelov, D. et al. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell 11, 1033–1041 (2003).
Doyen, C.M. et al. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol. Cell. Biol. 26, 1156–1164 (2006).
Agelopoulos, M. & Thanos, D. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J. 25, 4843–4853 (2006).
Ouararhni, K. et al. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev. 20, 3324–3336 (2006).
Nusinow, D.A. et al. Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J. Biol. Chem. 282, 12851–12859 (2007).
Karras, G.I. et al. The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920 (2005).
Kustatscher, G., Hothorn, M., Pugieux, C., Scheffzek, K. & Ladurner, A.G. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 12, 624–625 (2005).
Changolkar, L.N. & Pehrson, J.R. macroH2A1 histone variants are depleted on active genes but concentrated on the inactive X chromosome. Mol. Cell. Biol. 26, 4410–4420 (2006).
Eklund, E.A. The role of HOX genes in malignant myeloid disease. Curr. Opin. Hematol. 14, 85–89 (2007).
Houldsworth, J., Heath, S.C., Bosl, G.J., Studer, L. & Chaganti, R.S. Expression profiling of lineage differentiation in pluripotential human embryonal carcinoma cells. Cell Growth Differ. 13, 257–264 (2002).
Jørgensen, H.F. et al. Stem cells primed for action: Polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells. Cell Cycle 5, 1411–1414 (2006).
Squazzo, S.L. et al. SUZ12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 16, 890–900 (2006).
Boyer, L.A. et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 (2006).
Lee, T.I. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 (2006).
Bracken, A.P., Dietrich, N., Pasini, D., Hansen, K.H. & Helin, K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136 (2006).
Kuzmichev, A., Nishioka, K., Erdjument-Bromage, H., Tempst, P. & Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16, 2893–2905 (2002).
Kirmizis, A. et al. Silencing of human Polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 18, 1592–1605 (2004).
Leucht, C. et al. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat. Neurosci. 11, 641–648 (2008).
Changolkar, L.N. et al. Developmental changes in histone macroH2A1-mediated gene regulation. Mol. Cell. Biol. 27, 2758–2764 (2007).
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004).
Vicent, G.P. et al. DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol. Cell 16, 439–452 (2004).
Pasini, D., Bracken, A.P., Jensen, M.R., Lazzerini Denchi, E. & Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23, 4061–4071 (2004).
Buschbeck, M., Hofbauer, S., Croce, L.D., Keri, G. & Ullrich, A. Abl-kinase-sensitive levels of ERK5 and its intrinsic basal activity contribute to leukaemia cell survival. EMBO Rep. 6, 63–69 (2005).
Villa, R. et al. The methyl-CpG binding protein MBD1 is required for PML-RARα function. Proc. Natl. Acad. Sci. USA 103, 1400–1405 (2006).
Frank, S.R., Schroeder, M., Fernandez, P., Taubert, S. & Amati, B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15, 2069–2082 (2001).
O'Geen, H., Nicolet, C.M., Blahnik, K., Green, R. & Farnham, P.J. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques 41, 577–580 (2006).
Métivier, R. et al. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763 (2003).
Smyth, G.K. Limma: linear models for microarray data. in Bioinformatics and Computational Biology Solutions Using R and Bioconductor (Springer, New York, 2005).
Ritchie, M.E. et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23, 2700–2707 (2007).
Breitling, R., Armengaud, P., Amtmann, A. & Herzyk, P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573, 83–92 (2004).
Hong, F. et al. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22, 2825–2827 (2006).
Hosack, D.A., Dennis, G., Jr., Sherman, B.T., Lane, H.C. & Lempicki, R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 4, R70 (2003).
Dennis, G. Jr. et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4, 3 (2003).
Kimmel, C.B., Ballard, W.W., Kimmel, S.R., Ullmann, B. & Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995).
Thisse, C. & Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008).
Acknowledgements
We are indebted to T. Rasmussen, G. Dreyfuss (Howard Hughes Medical Institute) for the hnRNP U 4G5 antibody, S. Dimitrov and C. Pujades for reagents and members of the Di Croce laboratory for helpful discussions. This work was supported by grants from the Spanish “Ministerio de Educación y Ciencia” and from “La Marató TV3” and Consolider. M.B. was supported by a Fellowship from Deutsche Forschungsgesellschaft (DFG) and I.U. by a predoctoral fellowship from Spanish “Ministerio de Educación y Ciencia”.
Author information
Authors and Affiliations
Contributions
M.B., I.U., I.W., A.G. and L.M. performed the experiments; M.B., P.R., D.M., R.G., H.L.-S. and L.D.C. analyzed the data; M.B. and L.D.C. designed the project and wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1–3 (PDF 4174 kb)
Rights and permissions
About this article
Cite this article
Buschbeck, M., Uribesalgo, I., Wibowo, I. et al. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol 16, 1074–1079 (2009). https://doi.org/10.1038/nsmb.1665
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1665
This article is cited by
-
Histone macroH2A1 is a stronger regulator of hippocampal transcription and memory than macroH2A2 in mice
Communications Biology (2022)
-
The roles of histone variants in fine-tuning chromatin organization and function
Nature Reviews Molecular Cell Biology (2020)
-
LSH mediates gene repression through macroH2A deposition
Nature Communications (2020)
-
Histone variant MacroH2A1 is downregulated in prostate cancer and influences malignant cell phenotype
Cancer Cell International (2019)
-
DNA replication dynamics of vole genome and its epigenetic regulation
Epigenetics & Chromatin (2019)