Abstract
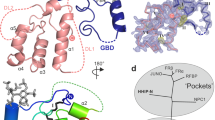
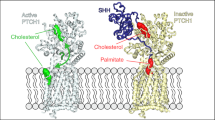
Hedgehog (Hh) signaling is crucial for many aspects of embryonic development, whereas dysregulation of this pathway is associated with several types of cancer. Hedgehog-interacting protein (Hhip) is a surface receptor antagonist that is equipotent against all three mammalian Hh homologs. The crystal structures of human HHIP alone and bound to Sonic hedgehog (SHH) now reveal that HHIP is comprised of two EGF domains and a six-bladed β-propeller domain. In the complex structure, a critical loop from HHIP binds the pseudo active site groove of SHH and directly coordinates its Zn2+ cation. Notably, sequence comparisons of this SHH binding loop with the Hh receptor Patched (Ptc1) ectodomains and HHIP- and PTC1-peptide binding studies suggest a 'patch for Patched' at the Shh pseudo active site; thus, we propose a role for Hhip as a structural decoy receptor for vertebrate Hh.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ingham, P.W. & McMahon, A.P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087 (2001).
Jiang, J. & Hui, C.C. Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812 (2008).
Beachy, P.A., Karhadkar, S.S. & Berman, D.M. Tissue repair and stem cell renewal in carcinogenesis. Nature 432, 324–331 (2004).
Scales, S.J. & de Sauvage, F.J. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol. Sci. 30, 303–312 (2009).
Lee, J.J. et al. Autoproteolysis in hedgehog protein biogenesis. Science 266, 1528–1537 (1994).
Porter, J.A. et al. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature 374, 363–366 (1995).
Porter, J.A. et al. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 86, 21–34 (1996).
Porter, J.A., Young, K.E. & Beachy, P.A. Cholesterol modification of hedgehog signaling proteins in animal development. Science 274, 255–259 (1996).
Pepinsky, R.B. et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 273, 14037–14045 (1998).
Fuse, N. et al. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for Patched. Proc. Natl. Acad. Sci. USA 96, 10992–10999 (1999).
Chen, Y. & Struhl, G. Dual roles for patched in sequestering and transducing Hedgehog. Cell 87, 553–563 (1996).
Marigo, V., Davey, R.A., Zuo, Y., Cunningham, J.M. & Tabin, C.J. Biochemical evidence that Patched is the Hedgehog receptor. Nature 384, 176–179 (1996).
Stone, D.M. et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129–134 (1996).
Rohatgi, R. & Scott, M.P. Patching the gaps in Hedgehog signalling. Nat. Cell Biol. 9, 1005–1009 (2007).
Lum, L. et al. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299, 2039–2045 (2003).
Yao, S., Lum, L. & Beachy, P. The Ihog cell-surface proteins bind Hedgehog and mediate pathway activation. Cell 125, 343–357 (2006).
Kang, J.S. et al. CDO: an oncogene-, serum-, and anchorage-regulated member of the Ig/fibronectin type III repeat family. J. Cell Biol. 138, 203–213 (1997).
Zhang, W., Kang, J.S., Cole, F., Yi, M.J. & Krauss, R.S. Cdo functions at multiple points in the Sonic Hedgehog pathway, and Cdo-deficient mice accurately model human holoprosencephaly. Dev. Cell 10, 657–665 (2006).
Kang, J.S., Mulieri, P.J., Hu, Y., Taliana, L. & Krauss, R.S. BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. EMBO J. 21, 114–124 (2002).
Tenzen, T. et al. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev. Cell 10, 647–656 (2006).
Allen, B.L., Tenzen, T. & McMahon, A.P. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 21, 1244–1257 (2007).
McLellan, J.S. et al. Structure of a heparin-dependent complex of Hedgehog and Ihog. Proc. Natl. Acad. Sci. USA 103, 17208–17213 (2006).
McLellan, J.S. et al. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature 455, 979–983 (2008).
Chuang, P.T. & McMahon, A.P. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature 397, 617–621 (1999).
Chuang, P.T., Kawcak, T. & McMahon, A.P. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 17, 342–347 (2003).
Olsen, C.L., Hsu, P.P., Glienke, J., Rubanyi, G.M. & Brooks, A.R. Hedgehog-interacting protein is highly expressed in endothelial cells but down-regulated during angiogenesis and in several human tumors. BMC Cancer 4, 43 (2004).
Pathi, S. et al. Comparative biological responses to human Sonic, Indian, and Desert hedgehog. Mech. Dev. 106, 107–117 (2001).
Frank-Kamenetsky, M. et al. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J. Biol. 1, 10 (2002).
Yauch, R.L. et al. A paracrine requirement for hedgehog signalling in cancer. Nature 455, 406–410 (2008).
Jeon, H. et al. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat. Struct. Biol. 8, 499–504 (2001).
Oubrie, A. Structure and mechanism of soluble glucose dehydrogenase and other PQQ-dependent enzymes. Biochim. Biophys. Acta 1647, 143–151 (2003).
Southall, S.M., Doel, J.J., Richardson, D.J. & Oubrie, A. Soluble aldose sugar dehydrogenase from Escherichia coli: a highly exposed active site conferring broad substrate specificity. J. Biol. Chem. 281, 30650–30659 (2006).
Oubrie, A., Rozeboom, H.J. & Dijkstra, B.W. Active-site structure of the soluble quinoprotein glucose dehydrogenase complexed with methylhydrazine: a covalent cofactor-inhibitor complex. Proc. Natl. Acad. Sci. USA 96, 11787–11791 (1999).
Hall, T.M., Porter, J.A., Beachy, P.A. & Leahy, D.J. A potential catalytic site revealed by the 1.7-Å crystal structure of the amino-terminal signalling domain of Sonic hedgehog. Nature 378, 212–216 (1995).
Briscoe, J., Chen, Y., Jessell, T.M. & Struhl, G. A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Mol. Cell 7, 1279–1291 (2001).
Taipale, J., Cooper, M.K., Maiti, T. & Beachy, P.A. Patched acts catalytically to suppress the activity of Smoothened. Nature 418, 892–897 (2002).
Ericson, J., Morton, S., Kawakami, A., Roelink, H. & Jessell, T.M. Two critical periods of Sonic hedgehog signaling required for the specification of motor neuron identity. Cell 87, 661–673 (1996).
Rawlings, N.D., Morton, F.R., Kok, C.Y., Kong, J. & Barrett, A.J. MEROPS: the peptidase database. Nucleic Acids Res. 36, D320–D325 (2008).
Bochtler, M., Odintsov, S.G., Marcyjaniak, M. & Sabala, I. Similar active sites in lysostaphins and D-Ala-D-Ala metallopeptidases. Protein Sci. 13, 854–861 (2004).
Bussiere, D.E. et al. The structure of VanX reveals a novel amino-dipeptidase involved in mediating transposon-based vancomycin resistance. Mol. Cell 2, 75–84 (1998).
Gomis-Rüth, F.X. et al. Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1. Nature 389, 77–81 (1997).
Stamos, J., Lazarus, R.A., Yao, X., Kirchhofer, D. & Wiesmann, C. Crystal structure of the HGF β-chain in complex with the Sema domain of the Met receptor. EMBO J. 23, 2325–2335 (2004).
Kirchhofer, D. et al. Structural and functional basis of the serine protease-like hepatocyte growth factor β-chain in Met binding and signaling. J. Biol. Chem. 279, 39915–39924 (2004).
Kawahira, H. et al. Combined activities of hedgehog signaling inhibitors regulate pancreas development. Development 130, 4871–4879 (2003).
Dubourg, C. et al. Molecular screening of SHH, ZIC2, SIX3, and TGIF genes in patients with features of holoprosencephaly spectrum: mutation review and genotype-phenotype correlations. Hum. Mutat. 24, 43–51 (2004).
Gao, B. et al. Mutations in IHH, encoding Indian hedgehog, cause brachydactyly type A-1. Nat. Genet. 28, 386–388 (2001).
McCready, M.E. et al. A novel mutation in the IHH gene causes brachydactyly type A1: a 95-year-old mystery resolved. Hum. Genet. 111, 368–375 (2002).
Kirkpatrick, T.J. et al. Identification of a mutation in the Indian Hedgehog (IHH) gene causing brachydactyly type A1 and evidence for a third locus. J. Med. Genet. 40, 42–44 (2003).
Gao, B. & He, L. Answering a century old riddle: Brachydactyly type A1. Cell Res. 14, 179–187 (2004).
Liu, M. et al. A novel heterozygous mutation in the Indian hedgehog gene (IHH) is associated with brachydactyly type A1 in a Chinese family. J. Hum. Genet. 51, 727–731 (2006).
Gao, B. et al. A mutation in Ihh that causes digit abnormalities alters its signalling capacity and range. Nature 458, 1196–1200 (2009).
Byrnes, A.M. et al. Brachydactyly A-1 mutations restricted to the central region of the N-terminal active fragment of Indian hedgehog. Eur. J. Hum. Genet. published online, doi:10.1038/ejhg.2009.18 (11 March 2009).
St-Jacques, B., Hammerschmidt, M. & McMahon, A.P. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 13, 2072–2086 (1999).
Kiselyov, A.S., Tkachenko, S.E., Balakin, K.V. & Ivachtchenko, A.V. Small-molecule modulators of Hh and Wnt signaling pathways. Expert Opin. Ther. Targets 11, 1087–1101 (2007).
Stanton, B.Z. et al. A small molecule that binds Hedgehog and blocks its signaling in human cells. Nat. Chem. Biol. 5, 154–156 (2009).
Pepinsky, R.B. et al. Mapping sonic hedgehog-receptor interactions by steric interference. J. Biol. Chem. 275, 10995–11001 (2000).
Carpenter, D. et al. Characterization of two Patched receptors for the vertebrate Hedgehog protein family. Proc. Natl. Acad. Sci. USA 95, 13630–13634 (1998).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 (2003).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C. & Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 (2005).
Winn, M.D., Isupov, M.N. & Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 (2001).
Acknowledgements
We thank our colleagues H. Tian, T. Tang, F. Rousseau, C. Noguère, A. Esteves and C. Quan for reagents and technical advice; D. Arnott and E. Drake for 5E1 mapping and affinities; A. Spura and D. Dayne at ForteBio for their support; S. Fakra at the Advanced Light Source (ALS) for her assistance with calcium identification; and our collaborators at Curis, Inc. for S12 cells. Stanford Synchrotron Radiation Laboratory (SSRL), ALS and the Berkeley Center for Structural Biology are supported by the US Department of Energy, National Institutes of Health and the National Institute of General Medical Sciences.
Author information
Authors and Affiliations
Contributions
I.B. performed structural biology; H.R.M. expressed and purified proteins and performed biochemical assays; S.J.S. and X.W. performed cell-based assays; A.L. contributed to NMR experiments; J.F.B. carried out computational analysis of the Hhip ECD; I.B., H.R.M., S.J.S., A.L., J.F.B., F.J.d.S., S.G.H. and R.A.L. analyzed data and wrote the manuscript. All authors are employees of Genentech, Inc.
Corresponding authors
Ethics declarations
Competing interests
All authors are employees of Genentech, Inc.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Table 1 and Supplementary Methods (PDF 1223 kb)
Rights and permissions
About this article
Cite this article
Bosanac, I., Maun, H., Scales, S. et al. The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nat Struct Mol Biol 16, 691–697 (2009). https://doi.org/10.1038/nsmb.1632
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1632
This article is cited by
-
Suppression of apoptosis impairs phalangeal joint formation in the pathogenesis of brachydactyly type A1
Nature Communications (2024)
-
Hedgehog signaling in tissue homeostasis, cancers, and targeted therapies
Signal Transduction and Targeted Therapy (2023)
-
Therapeutic implication of Sonic Hedgehog as a potential modulator in ischemic injury
Pharmacological Reports (2023)
-
Hedgehog interacting protein activates sodium–glucose cotransporter 2 expression and promotes renal tubular epithelial cell senescence in a mouse model of type 1 diabetes
Diabetologia (2023)
-
Association of Sonic Hedgehog with the extracellular matrix requires its zinc-coordination center
BMC Molecular and Cell Biology (2021)